Differential Analysis
1. Identify Genomic Regions With Significantly Different Signal Between Conditions
The limma_column_cluster_differential_regions() function
uses the limma-voom pipeline to identify genomic regions with
significantly different signal between conditions (e.g., treatment
vs. control).
What this function does: - Compares two experimental conditions with biological replicates - Identifies significantly different genomic regions - Supports optional feature clustering - Applies multiple filtering thresholds - Generates quality control visualizations
Parameters
| Parameter | Type | Required | Description | Example |
|---|---|---|---|---|
sample_names |
character vector | Yes | Sample names (condition1 first, then condition2) | sample_names = c("V1", "V2", "T1", "T2") |
case_num |
integer | Yes | Number of replicates in condition 1 (≥2) | case_num = 2 |
control_num |
integer | Yes | Number of replicates in condition 2 (≥2) | control_num = 2 |
wgc_file_path |
character vector | Yes | Paths to count matrix files (.feather format) | wgc_file_path = c(path1, path2, path3, path4) |
sig_result_dir |
character | Yes | Output directory path | sig_result_dir = "output/differential_1" |
col_cluster_file |
character | No | Path to column cluster tsv file (default: NULL, treat each col as a cluster) | col_cluster_file = "clusters.tsv" |
normalization_factor |
numeric | No | CPM scaling factor (default: 1E6) | |
lowess_span |
numeric | No | Voom lowess span (default: 0.5) | |
l2fc_thres |
numeric | No | Log2 fold change threshold (default: 0.5) | |
mean_per_thres_list |
numeric vector | No | Expression percentile thresholds (default: 0.25) | |
fdr_thres_list |
numeric vector | No | FDR thresholds (default: 0.25) | |
pseudocount_for_log |
numeric | No | Pseudocount for log2 transformation (default: 1) |
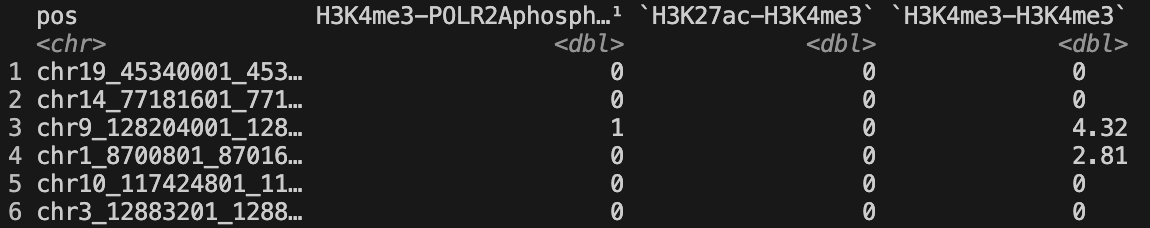
Required columns for col_cluster_file:
- feature: Samples: “H3K27me3-H3K27me3”, “H3K4me3-H3K4me3”,
… - label: Labels: 1,2,3…
Output Files
The function generates several output files:
- Voom plots -
voom_plot_lowess_span-{lowess_span}_post-filter-one_condition_nonzero-2_column_cluster-{col_label}.pdf- Quality control visualization showing mean-variance relationship before filtering
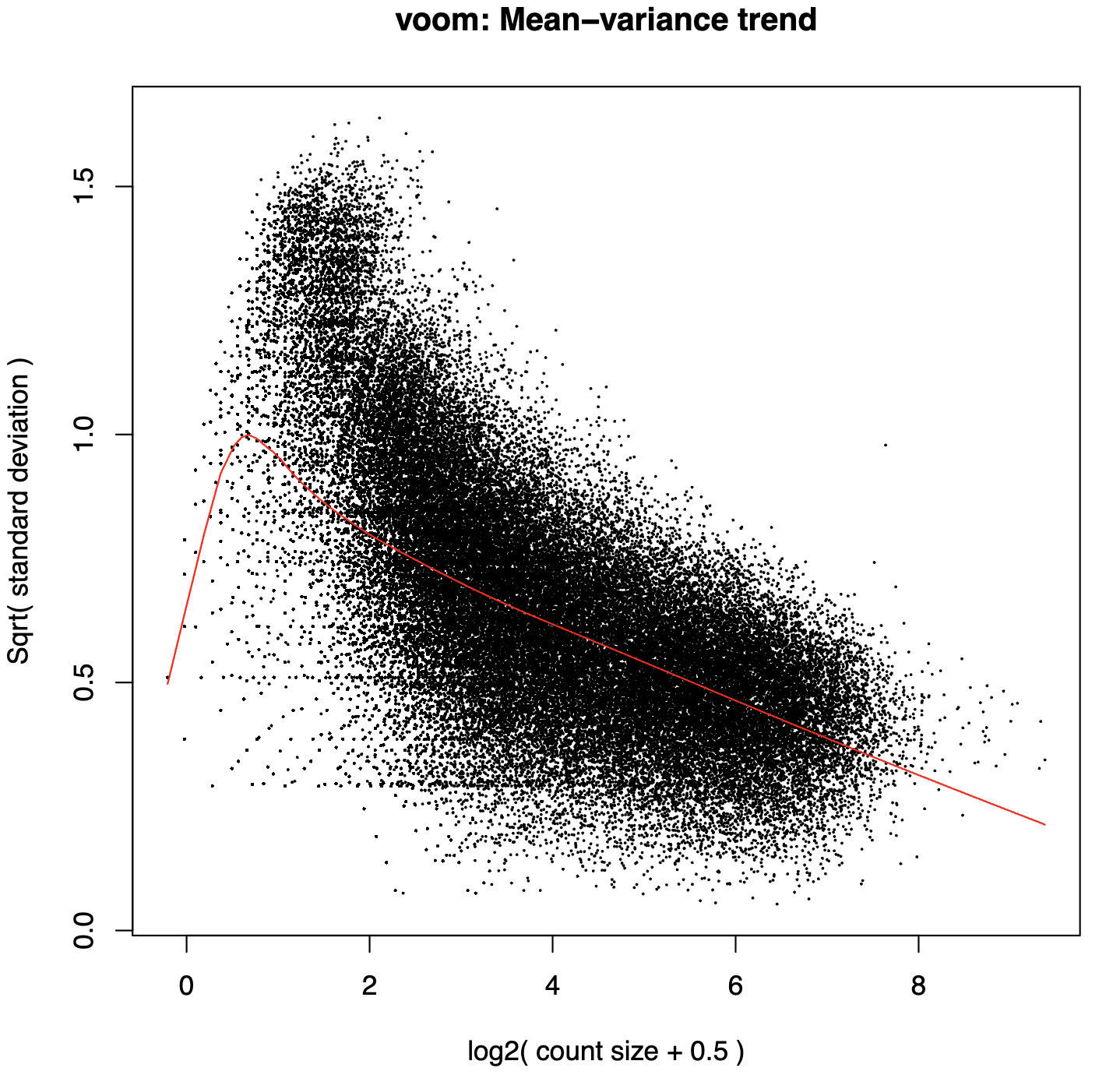
- Filtered voom plots -
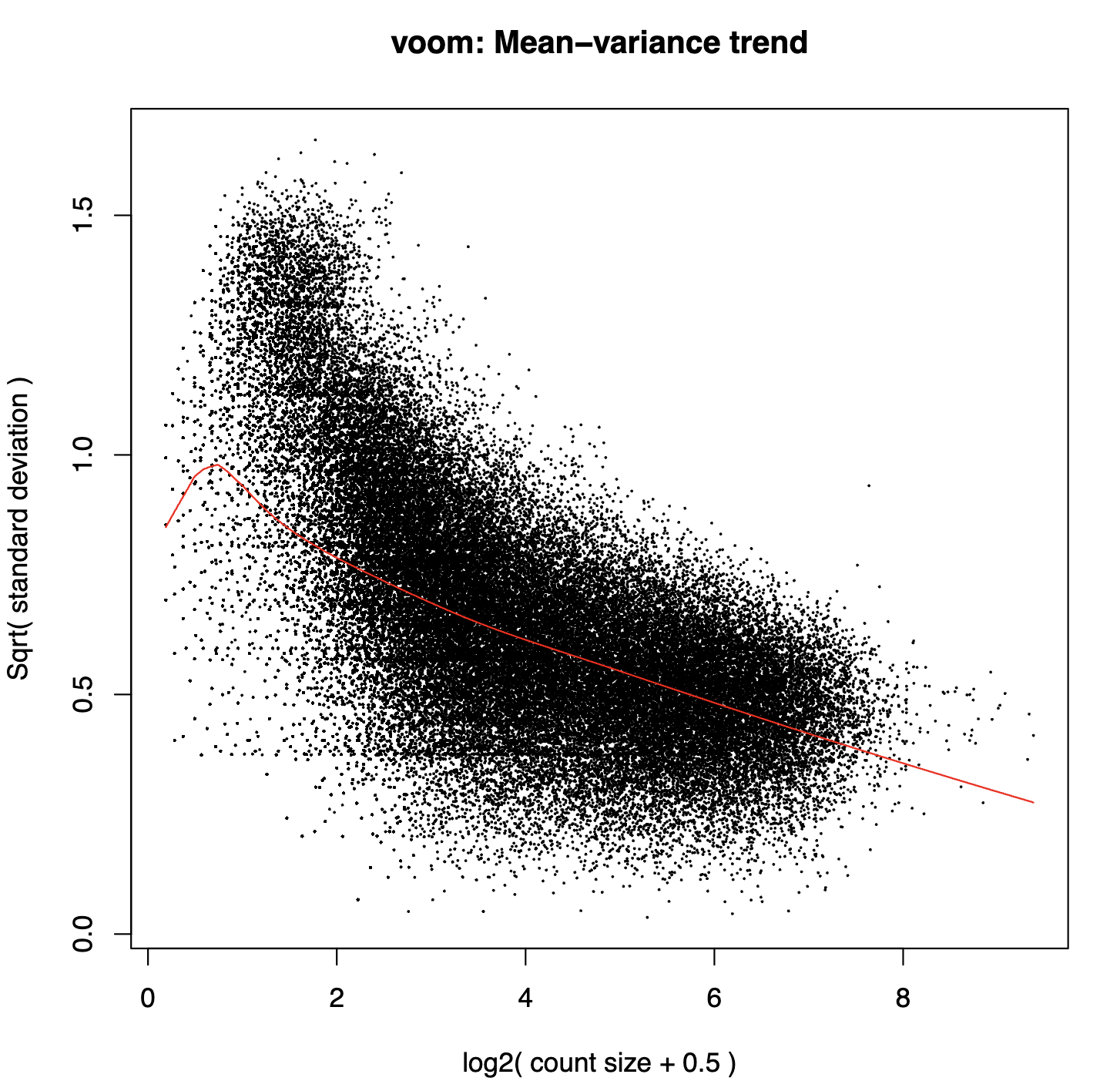
voom_plot_lowess_span-{lowess_span}_post-filter-one_condition_nonzero-2_rowmean-{mean_per_thres}_column_cluster-{col_label}.pdf- Voom plots after expression filtering
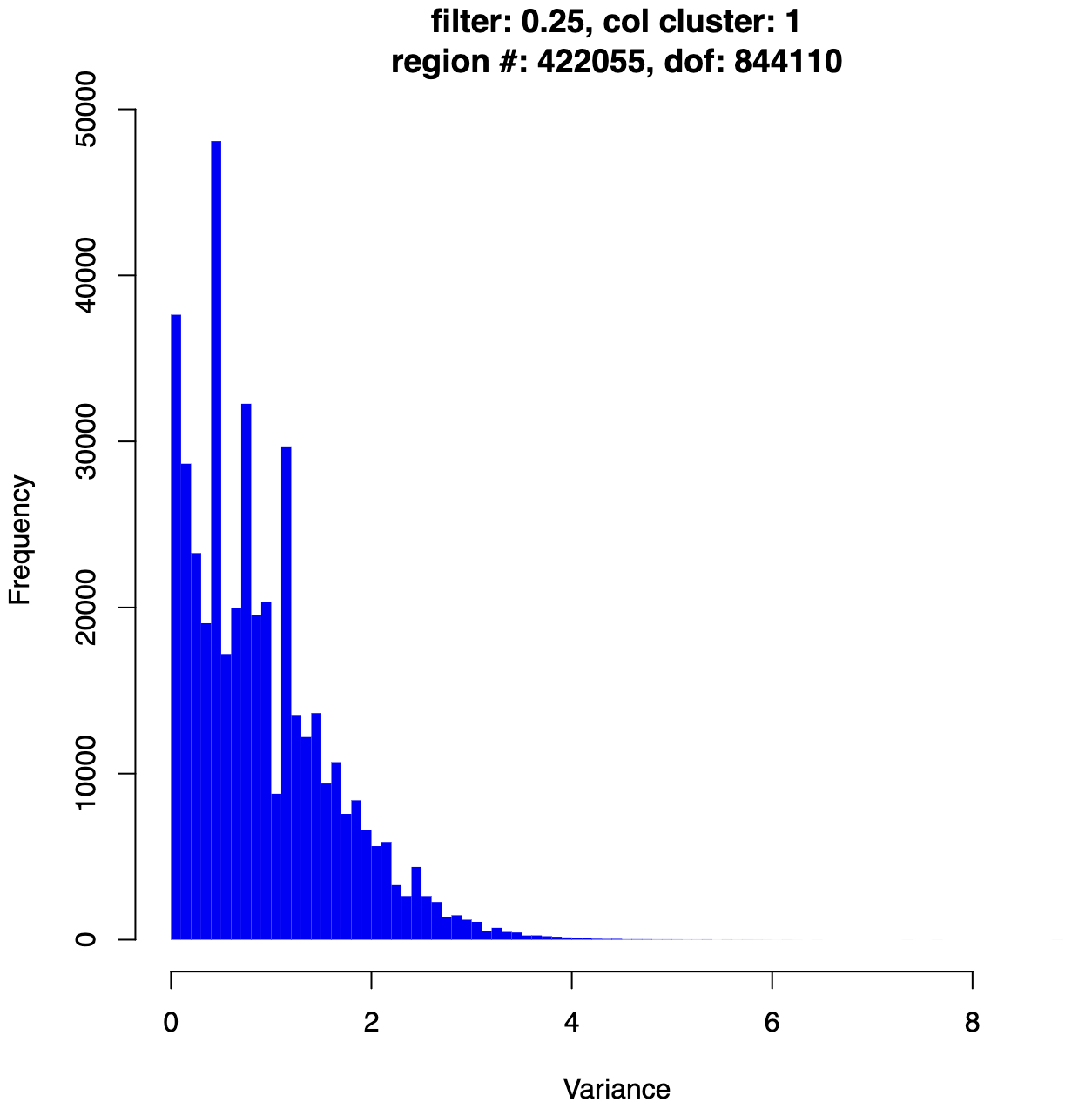
- Variance histograms -
hist_post-limmanorm_post-filter-one_condition_nonzero-2_rowmean-{mean_per_thres}_column_cluster-{col_label}_for_limma.pdf- Distribution of variance across regions after filtering
- All regions -
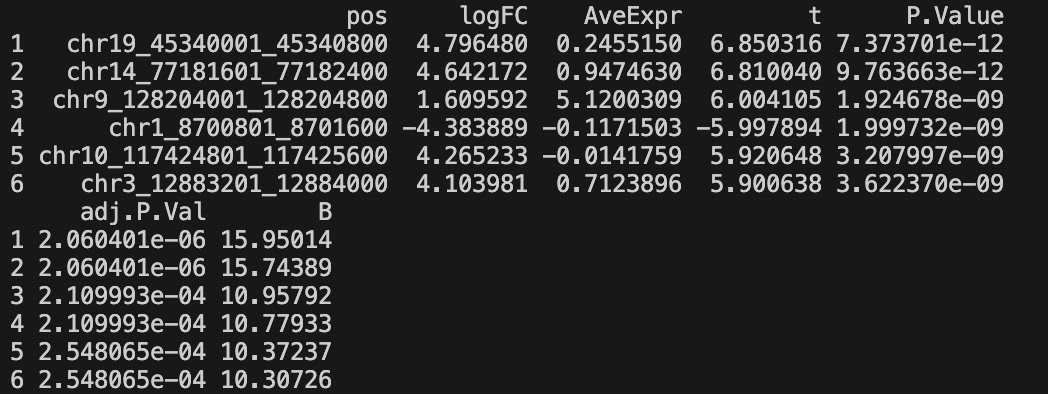
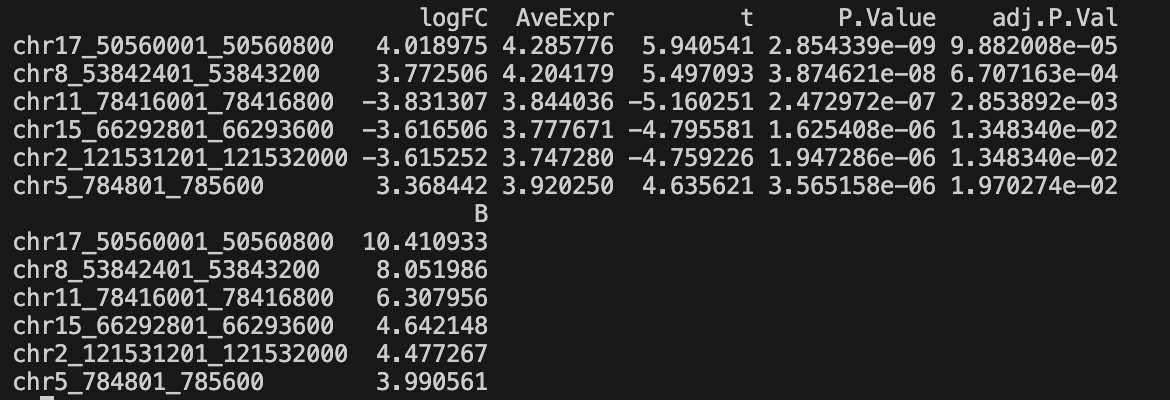
result_post-limmanorm_post-filter-one_condition_nonzero-2_rowmean-{mean_per_thres}_column_cluster-{col_label}_limma.feather- Complete results with statistics for all tested regions
- Columns: pos, logFC, AveExpr, t, P.Value, adj.P.Val, B
- Significant regions -
result_post-limmanorm_post-filter-one_condition_nonzero-2_rowmean-{mean_per_thres}_column_cluster-{col_label}_limma_FDR.tsv- Filtered results containing only significant regions (FDR < threshold & |logFC| > threshold)
- Log2 transformed WGC files -
{sample_name}_post-limmanorm_post-filter-one_condition_nonzero-2_rowmean-{mean_per_thres}_log2_column_cluster-{col_label}_limma_FDR-{fdr_thres}_logFC-{l2fc_thres}.feather- Log2-transformed count matrices for significant regions only
- One file per sample containing all features in the cluster
Example Usage
library(multiEpiCore)
sample_names = c("V1", "V2", "T1", "T2")
wgc_file_path = c(path1, path2, path3, path4)
sig_result_dir = "output/differential_1"
col_cluster_file = "input/clusters.tsv"
limma_column_cluster_differential_regions(
sample_names,
case_num = 2,
control_num = 2,
wgc_file_path,
sig_result_dir,
col_cluster_file
)2. Differential Expression Heatmaps
The plot_diff_heatmaps() function generate differential
expression heatmaps for multiple sample clusters, visualizing log2
expression patterns across conditions with customizable formatting and
clustering options.
What this function does:
- Creates heatmaps for differential expression visualization
- Organizes features by cluster assignments
- Displays expression patterns across sample groups
- Uses blue-white-red color scheme for log2 values
- Supports customizable formatting and layout options
- Generates publication-ready PDF outputs
Parameters
| Parameter | Type | Required | Description | Example |
|---|---|---|---|---|
sample_names |
character vector | Yes | Sample names (displayed as column titles) | sample_names = c("V1", "V2", "T1", "T2") |
sig_result_dir |
character | Yes | Output directory for heatmap PDFs | sig_result_dir = "output/differential_2" |
load_dir |
character | Yes | Directory containing expression matrix feather files | wgc_file_dir = "output/differential_1" |
col_cluster_file |
character | No | Path to column cluster tsv file (default: NULL, treat each col as a cluster) | col_cluster_file = "clusters.tsv" |
cluster_idx_list |
integer vector | No | Cluster indices to process (default: all cluster) | cluster_idx_list = c(1, 2, 3) |
mean_per_thres |
character | No | Mean expression percentile threshold (default: “0.25”) | mean_per_thres = "0.25" |
fdr_thres |
character | No | FDR threshold (default: “0.25”) | fdr_thres = "0.25" |
l2fc_thres |
numeric | No | Log2 fold change threshold (default: 0.5) | l2fc_thres = 0.5 |
show_heatmap_legend |
character | No | Display heatmap legend: “on”/“off” (default: “off”) | show_heatmap_legend = "on" |
show_colnames |
character | No | Display column names: “on”/“off” (default: “off”) | show_colnames = "on" |
col_size_coef |
numeric | No | Column width adjustment coefficient (default: 20) | col_size_coef = 15 |
colnames_fontsize |
numeric | No | Font size for column names (default: 10) | colnames_fontsize = 8 |
width_base |
numeric | No | Base width in mm for sizing (default: 8) | width_base = 10 |
random_seed |
integer | No | Random seed for reproducible rendering (default: 42) | random_seed = 123 |
font_size |
numeric | No | Font size for text elements (default: 50) | font_size = 40 |
target_pair_mapping_df_path |
character | No | Path to target name mapping file (NULL for no mapping) | target_pair_mapping_df_path = "mapping.tsv" |
Required columns for col_cluster_file:
- feature: Samples: “H3K27me3-H3K27me3”, “H3K4me3-H3K4me3”,
… - label: Labels: 1,2,3…
Output Files
The function generates PDF heatmap files:
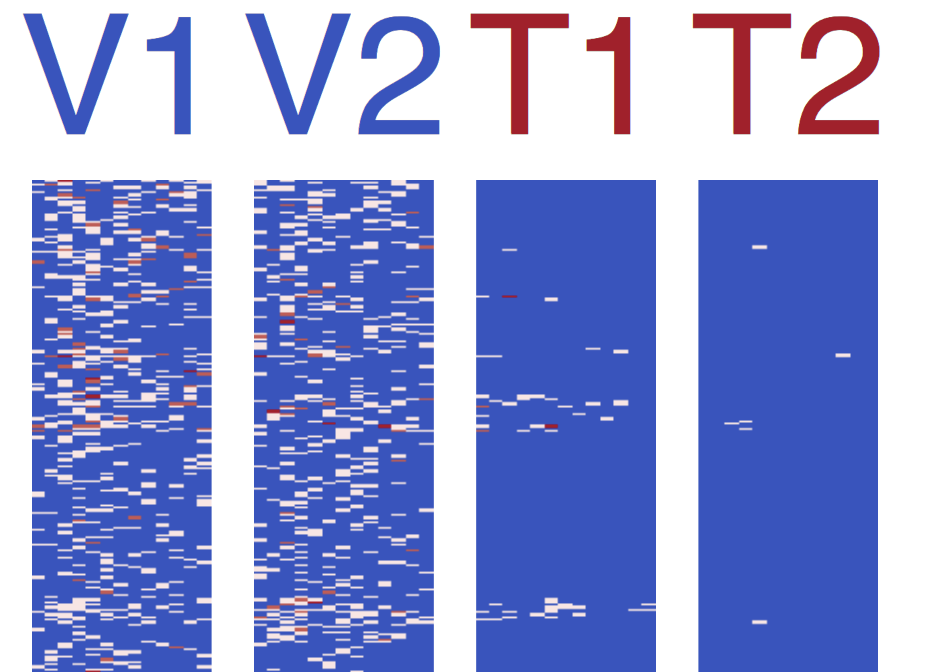
- Heatmaps -
diff_heatmap_col_cluster-{cluster_idx}_colname-{show_colnames}_col-reorder_size-{col_size_coef}.pdf- Heatmap with blue-white-red color gradient for log2 expression values
- Rows represent genomic regions, columns represent features
- Column titles color-coded by condition (blue for condition1, red for condition2)
Example Usage
library(multiEpiCore)
sample_names = c("V1", "V2", "T1", "T2")
sig_result_dir = "output/differential_2"
load_dir = "output/differential_1"
col_cluster_file = "input/clusters.tsv"
plot_diff_heatmaps(sample_names, sig_result_dir = sig_result_dir, load_dir = load_dir, col_cluster_file = col_cluster_file, cluster_idx_list = c(1:8, 10:15))3. Scatter Plots Comparing log2 Fold Changes
The generate_cluster_scatter_plots() function compares
log2 fold changes between genomic bins and RNA-seq expression for genes
within a specified distance, performing Fisher’s exact test to assess
concordance between epigenetic and transcriptomic changes across
clusters.
What this function does:
Maps genomic bins to gene promoters within a distance cutoff
Compares epigenetic changes (genomic bins) with transcriptomic changes (RNA-seq)
Generates scatter plots with density coloring for significant regions
Performs quadrant analysis to identify concordant/discordant changes
Conducts Fisher’s exact test to assess overall concordance
Creates publication-ready visualizations with statistical annotations
Parameters
| Parameter | Type | Required | Description | Example |
|---|---|---|---|---|
rnaseq_file_path |
character | Yes | Path to CSV file with RNA-seq differential expression results | rnaseq_file_path = "rnaseq_results.csv" |
promoter_file_path |
character | Yes | Path to TSV file with promoter annotations | promoter_file_path = "promoters.tsv" |
load_dir |
character | Yes | Directory containing limma results feather files | load_dir = "output/differential_1" |
sig_result_dir |
character | Yes | Output directory for plots and tables | sig_result_dir = "output/differential_3" |
cluster_idx_list |
integer vector | No | Cluster IDs to process (default: NULL = all clusters) | cluster_idx_list = c(1, 2, 3) |
nearest_dist_cutoff |
integer | No | Maximum distance in bp for bin-promoter mapping (default: 5000) | nearest_dist_cutoff = 5000 |
l2fc_thres |
numeric | No | Log2 fold change threshold for significance (default: 0.5) | l2fc_thres = 0.5 |
ylim_top |
numeric | No | Upper y-axis limit for RNA-seq log2FC (default: 7) | ylim_top = 7 |
ylim_bottom |
numeric | No | Lower y-axis limit for RNA-seq log2FC (default: -4) | ylim_bottom = -4 |
xlim_top |
numeric | No | Upper x-axis limit for genomic bin logFC (default: 3) | xlim_top = 3 |
xlim_bottom |
numeric | No | Lower x-axis limit for genomic bin logFC (default: -3) | xlim_bottom = -3 |
Output Files
The function generates two types of output files for each cluster:
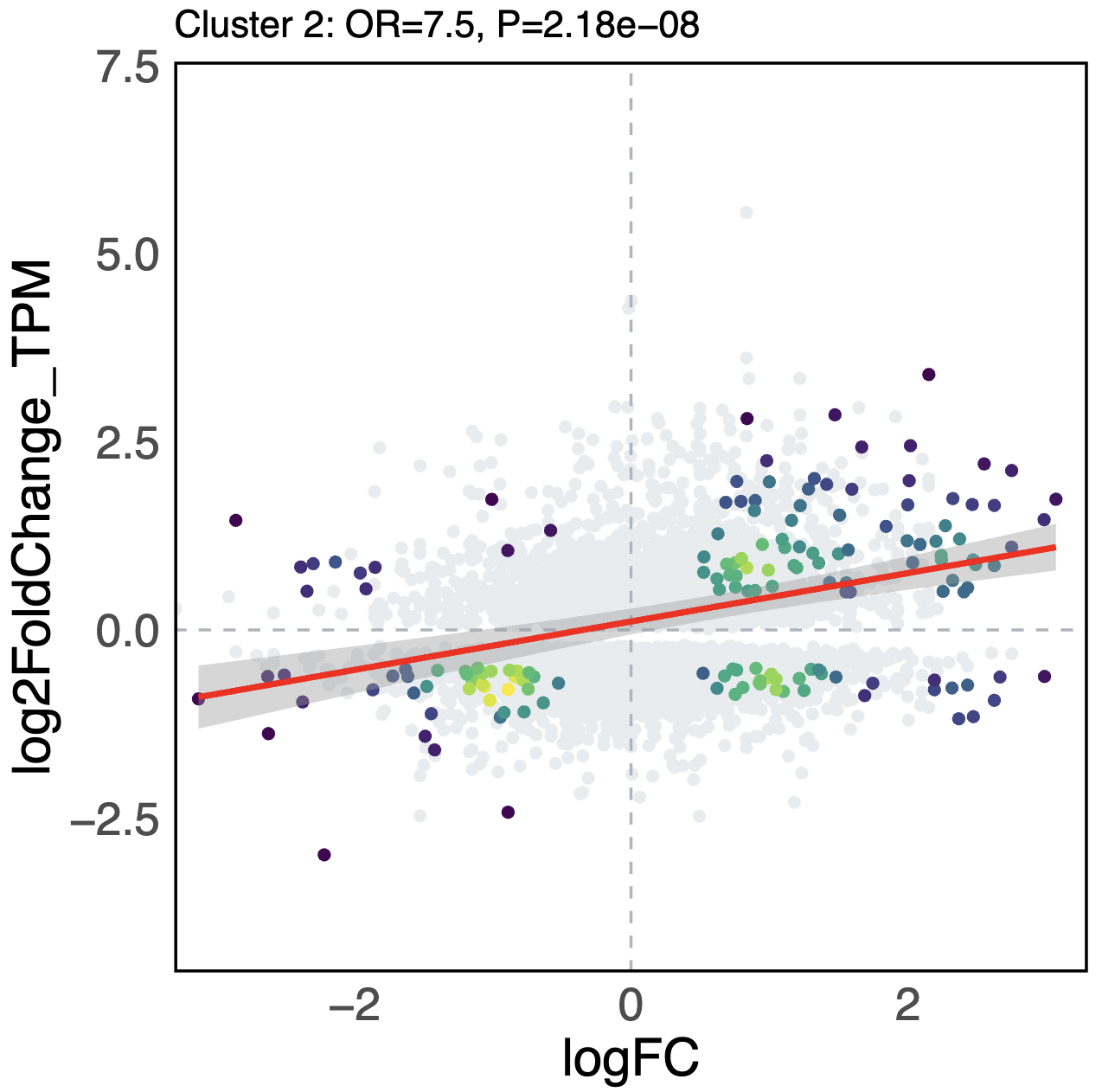
- Scatter plots -
scatter_{cluster_id}_{distance}_l2fc-{threshold}.pdf- Scatter plot comparing genomic bin log2FC (x-axis) vs RNA-seq log2FC (y-axis)
- Density coloring for significant points (viridis color scale)
- Grey points for non-significant regions
- Red regression line with confidence interval
- Title shows Fisher’s exact test results (Odds Ratio and P-value)
- Summary tables -
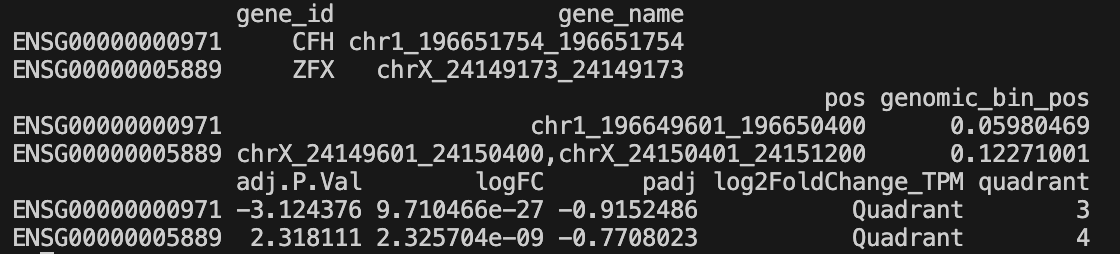
result_merge_bins_column_cluster-{cluster_id}_{distance}_l2fc-{threshold}.tsv- TSV file containing quadrant analysis results
- Columns: gene_id, gene_name, pos, genomic_bin_pos, adj.P.Val, logFC, padj, log2FoldChange_TPM, quadrant
- Only includes genes with significant changes
- Quadrants indicate concordance:
- Quadrant 1: Both up (genomic bin ↑, RNA-seq ↑) - Concordant
- Quadrant 2: Discordant (genomic bin ↓, RNA-seq ↑)
- Quadrant 3: Both down (genomic bin ↓, RNA-seq ↓) - Concordant
- Quadrant 4: Discordant (genomic bin ↑, RNA-seq ↓)
Example Usage
library(multiEpiCore)
rnaseq_file_path = "diff_expr_all_update.csv"
promoter_file_path = "promoters.tsv"
load_dir = "output/differential_1"
save_dir = "output/differential_3"
generate_cluster_scatter_plots(rnaseq_file_path, promoter_file_path, load_dir, save_dir, cluster_idx_list = c(1:8, 10:15))4. Summarize Significant Regions
The summary_sig_num() function summarizes the number of
significant differentially accessible regions by reading pre-computed
differential analysis results from
limma_column_cluster_differential_regions(), providing
counts of total, upregulated, and downregulated regions for each cluster
and threshold combination.
What this function does:
Reads pre-computed differential analysis results
Summarizes significant regions across multiple column clusters
Tests multiple filtering thresholds simultaneously
Generates summary tables with region counts (total, up, down)
Efficiently processes results without re-running analysis
Provides comprehensive overview of differential accessibility
Parameters
| Parameter | Type | Required | Description | Example |
|---|---|---|---|---|
load_dir |
character | Yes | Directory containing differential analysis results from limma_column_cluster_differential_regions | load_dir = "output/differential_1" |
sig_result_dir |
character | Yes | Output directory for summary tables | sig_result_dir = "output/differential_4" |
cluster_idx_list |
integer vector | Yes | Vector of column cluster labels to summarize | cluster_idx_list = c(1:8, 10:15) |
l2fc_thres |
numeric | No | Log2 fold change threshold (default: 0.5) | l2fc_thres = 0.5 |
mean_per_thres_list |
numeric vector | No | Expression percentile thresholds (default: 0.25) | mean_per_thres_list = c(0.25, 0.5) |
fdr_thres_list |
numeric vector | No | FDR thresholds (default: 0.25) | fdr_thres_list = c(0.05, 0.1) |
Output Files
The function generates summary TSV tables for each parameter combination:
- Summary tables -
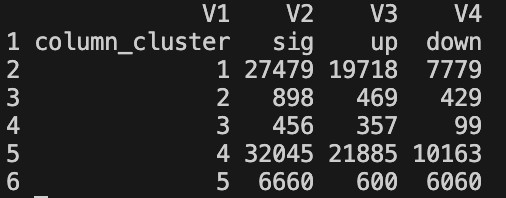
summary_sig_num_{mean_threshold}_FDR-{fdr_threshold}_log2FC-{l2fc_threshold}.tsv- TSV file with one row per column cluster
- Columns:
column_cluster: Cluster IDsig: Total number of significant regions (FDR < threshold AND |log2FC| > threshold)up: Number of upregulated regions (FDR < threshold AND log2FC > 0)down: Number of downregulated regions (FDR < threshold AND log2FC ≤ 0)
Example Usage
library(multiEpiCore)
load_dir = "output/differential_1"
sig_result_dir = "output/differential_4"
cluster_idx_list = c(1:8, 10:15)
summary_sig_num(load_dir = load_dir, sig_result_dir = sig_result_dir, cluster_idx_list = c(1:8, 10:15))5. Visualize Significant Region Counts
The summary_sig_num_log10_barplot_single_case() function
generates log10-scaled barplot visualization of significant region
counts, showing upregulated and downregulated regions across column
clusters with original count labels and pseudo-log transformation for
better visualization of wide dynamic ranges.
What this function does:
- Creates mirror barplots (up vs down) for each column cluster
- Applies log10 transformation for better visualization of wide ranges
- Uses pseudo-log scale to handle zero values gracefully
- Labels bars with original counts (not transformed values)
- Color-codes: upregulated (red/firebrick), downregulated (blue/steelblue)
- Generates publication-ready PDF outputs
Parameters
| Parameter | Type | Required | Description | Example |
|---|---|---|---|---|
result_file_dir |
character | Yes | Directory containing TSV file with summary statistics (from
summary_sig_num) |
result_file_dir = "output/differential_4" |
sig_result_dir |
character | Yes | Output directory for barplot PDF | sig_result_dir = "output/differential_5" |
mean_per_thres |
character | No | Mean expression percentile threshold (default: “0.25”) | mean_per_thres = "0.25" |
fdr_thres |
character | No | FDR threshold (default: “0.25”) | fdr_thres = "0.25" |
l2fc_thres |
numeric | No | Log2 fold change threshold (default: 0.5) | l2fc_thres = 0.5 |
Required columns in summary file:
column_cluster: Cluster IDsup: Number of upregulated regionsdown: Number of downregulated regions
Output Files
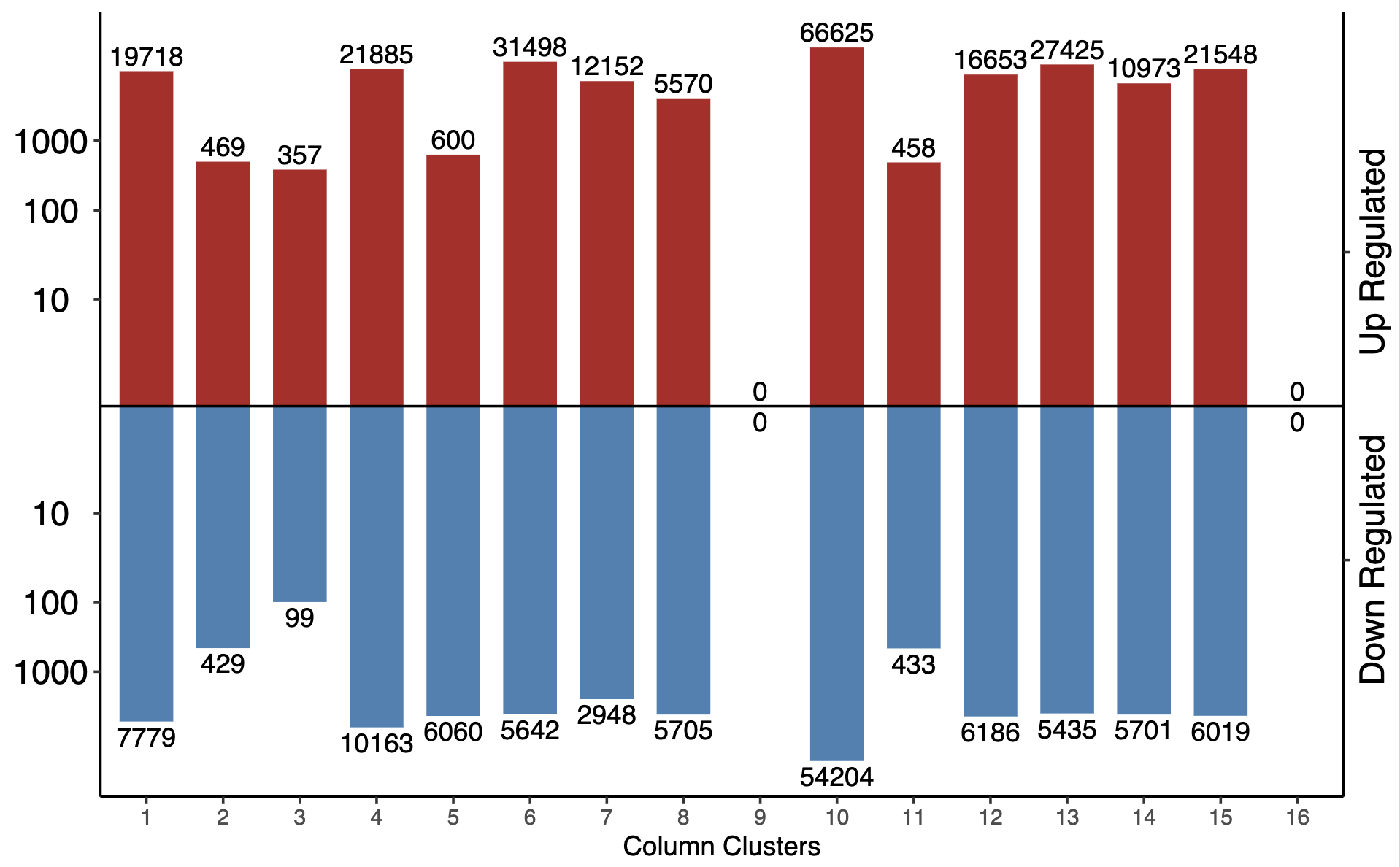
The function generates a PDF barplot:
- Barplot -
summary_sig_num_{mean_per_thres}_FDR-{fdr_thres}_log2FC-{l2fc_thres}_log.pdf- Mirror barplot with log10-transformed y-axis
- Upregulated regions: positive (red bars)
- Downregulated regions: negative (blue bars)
- Y-axis: pseudo-log10 scale with breaks at 10, 100, 1000
- Secondary y-axis: labels “Up Regulated” and “Down Regulated”
- Bar labels: original count values (not log-transformed)
- X-axis: column clusters
- Dimensions: 8” × 5”
Example Usage
library(multiEpiCore)
result_file_dir <- "output/differential_4"
sig_result_dir <- "output/differential_5"
summary_sig_num_log10_barplot_single_case(result_file_dir, sig_result_dir)