Biclustering Analysis
Wrapper Function
The biclustering_wrapper() function runs the full
biclustering analysis workflow in one call. It optionally merges
multiple input matrices, applies transformations, performs highly
variable region filtering, runs biclustering on the filtered matrix, and
generates downstream annotation results including genomic distribution
summaries and TFBS enrichment.
Parameters
| Parameter | Type | Default | Description | Example |
|---|---|---|---|---|
cm_path
|
character / character vector | — |
Path to the input count matrix in .feather format, or a
vector of paths to multiple matrices. When a vector is provided (length
> 1), matrices are merged before downstream processing.
|
cm_path = c(“cm1.feather”, “cm2.feather”)
|
out_dir
|
character | — | Output directory for all generated files, including transformed matrices, cluster tables, and annotation results. |
out_dir = “./biclustering_out”
|
apply_filter
|
logical |
TRUE
|
Whether to further filter genomic regions using
detect_hvr(). Recommended when the genome was segmented
into equal-sized bins (numeric regions). Set to
FALSE when the input matrix was built from user-provided
intervals (region file path), where additional filtering is typically
unnecessary.
|
apply_filter = TRUE
|
row_km
|
integer |
15
|
Number of k-means clusters for rows (genomic regions). |
row_km = 20
|
col_km
|
integer |
3
|
Number of k-means clusters for columns (CRF pairs). |
col_km = 4
|
apply_annotation
|
logical |
TRUE
|
Whether to perform downstream annotation on biclustered regions,
including genomic distribution summaries and TFBS enrichment.
Recommended for binned genomes (numeric regions). For
user-specified regions, set to FALSE if annotation is not
needed.
|
apply_annotation = TRUE
|
ref_genome
|
character |
“hg38”
|
Reference genome version used in annotation and control region
generation. Supported: “hg38”, “mm10”.
|
ref_genome = “mm10”
|
ref_source
|
character |
“knownGene”
|
Gene annotation source used in downstream analysis. Supported:
“knownGene” (UCSC knownGene via TxDb),
“GENCODE”.
|
ref_source = “GENCODE”
|
distributions
|
character vector |
c(“genic”,“ccre”)
|
Genomic feature distributions to summarize in the annotation step.
Options include: “genic”, “ccre”,
“cpg”, “repeat”.
|
distributions = c(“genic”,“repeat”)
|
plot
|
logical |
TRUE
|
Whether to generate diagnostic plots during filtering and biclustering
steps. This controls plotting behavior in detect_hvr() and
biclustering().
|
plot = FALSE
|
Example Usage
library(multiEpiCore)
# Test Data
cm_path <- c("count_matrix/C1_Count_Matrix_merged.feather",
"count_matrix/C2_Count_Matrix_merged.feather")
biclustering_wrapper(
cm_path = cm_path,
out_dir = "bicluster",
distributions = c("genic","ccre", "cpg", "repeat")
)1. Merge Count Matrices
The merge_count_matrices() function merges multiple
count matrices (Feather files) into a single count matrix by aligning
genomic regions and combining counts across samples.
Parameters
| Parameter | Type | Default | Description |
|---|---|---|---|
cm_path
|
character vector | - |
A vector of feather file paths to be merged (.feather)
|
out_dir
|
character |
“./”
|
Output directory |
check_consistency
|
boolean |
TRUE
|
If TRUE, only keep rows (positions) and columns (samples)
that exist in all input files. If FALSE, merge all
rows and columns, filling missing values with 0.
|
Example Usage
library(multiEpiCore)
# Test Data
cm_path <- c(
"count_matrix/C1_Count_Matrix_800.feather",
"count_matrix/C2_Count_Matrix_800.feather"
)
merge_count_matrices(cm_path = cm_path, out_dir = out_dir)2. Apply transformation
The apply_transformations() function performs a
sequential series of normalization and transformation steps on a count
matrix and outputs the processed result.
Parameters
| Parameter | Type | Default | Description |
|---|---|---|---|
cm_path
|
character | - |
Path to the input count matrix (.feather). The input file must be a
valid .feather file containing a positional column
(pos) or any first column that uniquely identifies genomic
intervals. All remaining columns must be numeric.
|
out_dir
|
character |
“./”
|
Output directory |
transformations
|
character vector |
c(“remove0”, “libnorm”, “log2p1”, “qnorm”)
|
Vector specifying the transformation pipeline. Steps are executed in order, and the behavior depends on the sequence you provide (see the table below for details). |
save_each_step
|
logical |
FALSE
|
If TRUE, writes a .feather file after each
step, allowing inspection of intermediate matrices.
|
The order of operations in transformations directly
determines the output. Below is the full list of supported steps:
| Transformation | Description |
|---|---|
remove0 |
Remove rows where all samples have zero counts |
libnorm |
Library size normalization (CPM, counts per million) |
log2p1 |
Log2 transformation: log2(x + 1) |
qnorm |
Quantile normalization across samples |
minmaxnorm |
Scale values to [0, 1] range |
sqrt |
Square root transformation |
Output Files
After processing all selected transformations, a final Feather file named:
<original_name>_transformed.featherIf set save_each_step = TRUE, the output path will
be:
out_dir/
├── [count_matrix_prefix]_transformed.feather
├── [count_matrix_prefix]_remove0.feather
├── [count_matrix_prefix]_remove0_libnorm.feather
├── [count_matrix_prefix]_remove0_libnorm_log2p1.feather
└── [count_matrix_prefix]_remove0_libnorm_log2p1_qnorm.featherExample Usage
library(multiEpiCore)
# Test Data
apply_transformations(
cm_path = "Count_Matrix_merged.feather",
transformations = c("remove0", "libnorm", "log2p1", "qnorm"),
out_dir = "bicluster",
save_each_step = FALSE
)3. Highly Variable Regions
This workflow provides two complementary approaches for identifying highly variable genomic regions:
Integrated workflow (recommended): The
detect_hvr()function combines mean-variance modeling and feature selection in a single step, internally callingmav_screen()andfilter_hvr()in sequence.Step-by-step workflow: For users who need fine-grained control or want to inspect intermediate results, the individual functions
mav_screen()andfilter_hvr()can be called separately.
Wrapper Function Overview
The detect_hvr() function streamlines the entire process
of identifying highly variable genomic regions by integrating
mean-variance modeling and feature selection into a single function
call.
Parameters
| Parameter | Type | Default | Description | Example |
|---|---|---|---|---|
transformed_cm_path
|
character | — |
Path to the transformed count matrix file in .feather
format. The file must contain a pos column.
|
transformed_cm_path = “data_transformed.feather”
|
out_dir
|
character |
“./”
|
Output directory path for all generated results. |
out_dir = “./hvr_results”
|
keep_percent
|
numeric |
0.01
|
Fraction of total regions to retain (range: 0–1). Selected regions are distributed equally across all bins. |
keep_percent = 0.05
|
log2mean_quantile_thres
|
numeric |
0.99
|
Quantile threshold (range: 0–1) applied to log2(mean) expression. Only regions above this threshold are retained in the final selection. |
log2mean_quantile_thres = 0.95
|
plot
|
logical |
FALSE
|
Whether to generate diagnostic plots for both MAV screening and feature selection steps. |
plot = TRUE
|
Example Usage
library(multiEpiCore)
# Test Data
path <- "bicluster/Count_Matrix_merged_transformed.feather"
detect_hvr(transformed_cm_path = path, out_dir = "./count_matrix/", plot = TRUE)3A. Model mean-variance relationship and calculate hypervariance metrics
The mav_screen() function models the mean–variance
relationship of transformed count data across genomic regions and
identifies regions exhibiting variability beyond technical expectations.
It computes hypervariance metrics that quantify each genomic region’s
deviation from the expected mean–variance trend.
What this function does:
(Model mean-variance relationship) Fits a regression model in log2 space to characterize the global relationship between mean signal intensity and variance across all genomic regions. This model captures the baseline mean–variance trend expected under technical variation.
(Calculate expected variance) For each genomic region, estimates the expected variance given its mean signal level using the fitted mean–variance model. This expected variance serves as the null reference for assessing excess variability.
(Normalize regional signals) Centers the signal of each genomic region by its observed mean and scales it by the expected standard deviation derived from the model. This normalization removes mean-dependent variance effects, enabling fair comparison of variability across regions with different signal levels.
(Quantify overdispersion) Computes hypervariance metrics for each genomic region, defined as the sum of squared normalized deviations divided by the corresponding degrees of freedom. Regions with elevated hypervariance values exhibit variability exceeding that expected from the global mean–variance relationship, consistent with biological heterogeneity rather than technical noise.
(Output region-level statistics) Outputs comprehensive region-level statistics, including observed mean and variance, expected variance, normalized values, and hypervariance metrics, to a feather file for downstream selection of highly variable genomic regions and quality control.
Parameters
| Parameter | Type | Default | Description | Example |
|---|---|---|---|---|
transformed_cm_path
|
character | — |
Path to the transformed count matrix file in .feather
format. The file must contain a pos column.
|
transformed_cm_path = “Count_Matrix_transformed.feather”
|
out_dir
|
character |
“./”
|
Output directory path for all generated results. |
out_dir = “./mav_screen_results”
|
fitting_model
|
character |
“gam”
|
Model used to fit the mean–variance relationship. Supported options are
“gam” and “loess”.
|
fitting_model = “loess”
|
k
|
integer |
40
|
Number of basis functions for GAM spline fitting. This parameter is only
used when fitting_model = “gam”.
|
k = 30
|
span
|
numeric |
0.5
|
Smoothing span for LOESS fitting. This parameter is only used when
fitting_model = “loess”.
|
span = 0.3
|
seed
|
integer |
42
|
Random seed used to ensure reproducibility during sampling. |
seed = 123
|
font_size
|
numeric |
10
|
Font size used for plot labels and axes. |
font_size = 12
|
nrow_sample_per
|
numeric |
0.2
|
Proportion of rows (range: 0–1) to randomly sample for density plot generation. |
nrow_sample_per = 0.1
|
plot
|
logical |
FALSE
|
Whether to generate and save diagnostic plots as .png
files.
|
plot = TRUE
|
Input data requirements:
The
transformed_cm_pathparameter refers directly to a pre-processed count matrix stored as a.featherfile. This file must not contain raw counts. Instead, the matrix should already be:- Pre-normalized (e.g., quantile normalization, TPM, RPKM, or other appropriate normalization)
- Log-transformed if necessary for your analysis
- Batch-corrected if applicable
- Quality-filtered to remove low-quality regions or samples
Additionally, the
.featherfile must have the first column named “pos” containing region identifiers (e.g., chr1_9601_10400).Model selection:
- Use GAM (default) for smooth, flexible fits with automatic complexity control
- Use LOESS for local fitting, better for non-linear patterns
Memory considerations: For very large datasets, adjust
nrow_sample_perto reduce memory usage for density plots.
Output Files
The function generates the following output files in the specified
out_dir (default: “./”):
- Mean-Variance Statistics File -
<input_name>_mav_screen.feather- Feather format dataframe with mean-variance statistics for each genomic region
- Contains the following columns:
pos: Original region identifiermean: Mean expression across all samplesvar: Variance across all samples
var_expect: Expected variance from fitted mean-variance modelnorm_mean: Mean of normalized values (should be ~0)norm_var: Variance of normalized values (should be ~1)hypervar: Hypervariance, sum of squared normalized values divided by (n-1)log2_mean: Log2-transformed mean expressionlog2_var: Log2-transformed variancelog2_var_expected: Log2-transformed expected variance from fitted model
pos mean var var_expect norm_mean norm_var hypervar chr1_9601_10400 0.60482747 3.2718638368 2.999402e+00 2.999402e+00 -2.400220e-17 1.0908388 chr1_10401_11200 0.06557750 0.1646858526 1.620011e-01 1.620011e-01 -6.149439e-18 1.0165723 chr1_12801_13600 0.03007577 0.0003917656 6.413126e-04 6.413126e-04 -1.840330e-18 0.6108808 chr1_14401_15200 0.02905376 0.0001423452 1.494525e-04 1.494525e-04 9.551020e-17 0.9524448 chr1_15201_16000 0.02994487 0.0007778492 5.396820e-04 5.396820e-04 -4.221334e-17 1.4413104 chr1_16001_16800 0.02875908 0.0001076822 9.580701e-05 9.580701e-05 -1.361187e-16 1.1239494 log2_mean log2_var log2_var_expected -0.7254044 1.710113 1.585175 -3.9306552 -2.602211 -2.625862 -5.0552546 -11.317722 -10.603762 -5.1051311 -12.778318 -12.731808 -5.0615474 -10.328222 -10.873420 -5.1198388 -13.180932 -13.352449 - Main diagnostic plots -
<input_name>_mean_variance.png(ifplot = TRUE)- Combined two-panel figure showing:
- Panel A: Log2(mean) vs Log2(variance) with fitted trend line in red
- Panel B: Log2(mean) vs Hypervariance
- Points shown with transparency to visualize density
- Theme: black and white with customizable font size
- Saved as PNG format
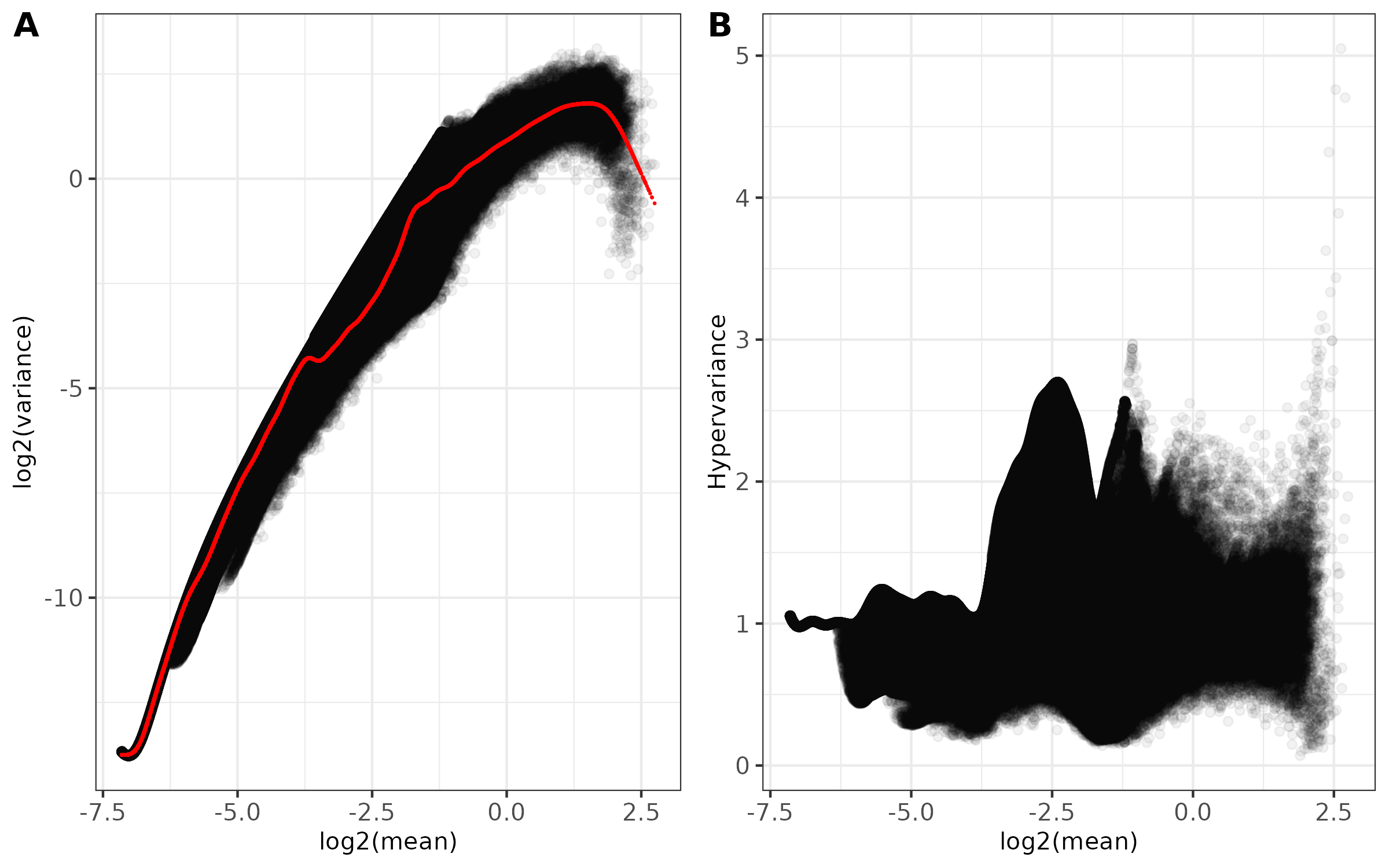
- Combined two-panel figure showing:
- Density plot -
<input_name>_fit_density.png(ifplot = TRUE)- Point density visualization of mean-variance relationship
- Uses viridis color scale to show point density
- Red points show expected variance from fitted model
- Based on randomly sampled subset of data (controlled by
nrow_sample_perandseed) - Useful for visualizing patterns in large datasets
- Saved as PNG format
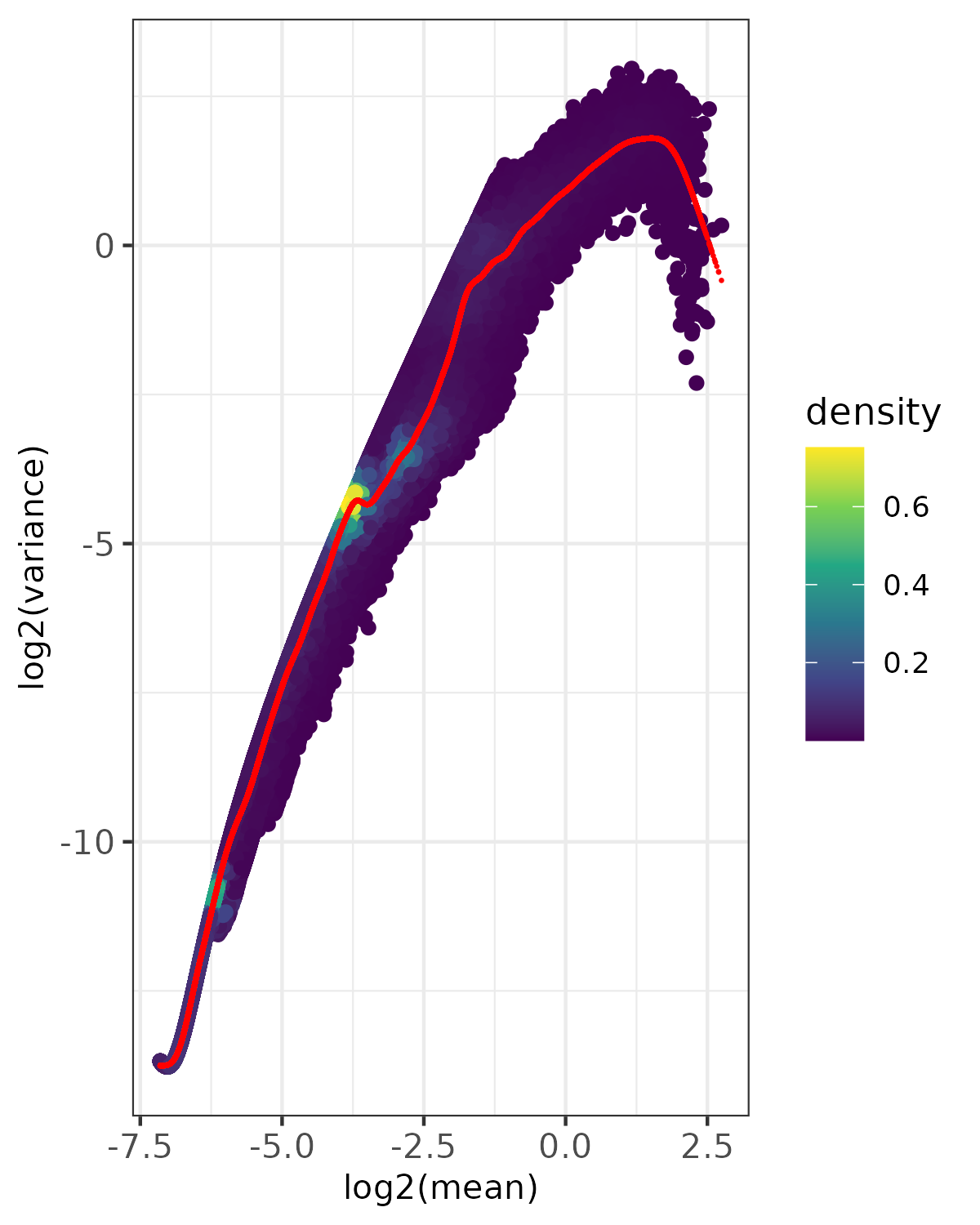
Example Usage
library(multiEpiCore)
# Test Data
trans_path <- "bicluster/Count_Matrix_merged_transformed.feather"
out_dir <- "bicluster"
mav_screen(transformed_cm_path = trans_path, out_dir = out_dir, plot=TRUE)3B. Select top-ranked regions based on hypervariance values
The filter_hvr() function identifies the most
informative genomic regions by combining hypervariance-based selection
with stratified sampling across expression levels.
What this function does:
(Stratified region selection) Divides the log2(mean) expression range into equal-sized bins to ensure balanced representation across low, medium, and high expression levels.
(Hypervariance filtering) Selects top hypervariant regions from each bin, prioritizing regions with biological variability (hypervariance > 1 indicates overdispersion).
(Expression threshold) Applies a percentile-based cutoff to retain only highly expressed regions, removing lowly expressed regions that may be noisy.
(Count matrix extraction) Retrieves corresponding count data for selected informative regions from the full transformed count matrix.
(Quality control visualization) Generates optional diagnostic plots showing:
- Mean-variance relationship with selected regions highlighted
- Hypervariance distribution across expression levels with selection boundaries
- Visual confirmation of balanced sampling strategy
(Efficient processing) Uses vectorized operations and selective column loading for fast processing of large genomic datasets.
Parameters
| Parameter | Type | Default | Description | Example |
|---|---|---|---|---|
transformed_cm_path
|
character | — |
Path to the transformed count matrix file in .feather
format. The file must contain a pos column as the region
identifier.
|
transformed_cm_path = “Count_Matrix_transformed.feather”
|
mav_stats_path
|
character | — |
Path to the mean–variance statistics file in .feather
format generated by the upstream MAV screening analysis.
|
mav_stats_path = “Count_Matrix_mav_screen.feather”
|
out_dir
|
character |
“./”
|
Output directory path used to save filtered results and optional diagnostic plots. |
out_dir = “./informative_results”
|
n_bins
|
integer |
100
|
Number of bins used to partition the log2(mean) range for stratified sampling, ensuring balanced representation across expression levels. |
n_bins = 50
|
keep_percent
|
numeric |
0.01
|
Fraction of total regions to retain (range: 0–1). Selected regions are distributed equally across all bins. |
keep_percent = 0.05
|
log2mean_quantile_thres
|
numeric |
0.99
|
Quantile threshold (range: 0–1) applied to log2(mean) expression. Only regions above this threshold are retained in the final selection. |
log2mean_quantile_thres = 0.95
|
plot
|
logical |
FALSE
|
Whether to generate diagnostic scatter plots visualizing the region selection process. |
plot = TRUE
|
Input file requirements:
mav_stats_path: Must be the output.featherfile frommav_screen()containing columns:pos,log2_mean,log2_var, andhypervar. The function will automatically select only these required columns during loading for memory efficiency.transformed_cm_path: Must be a pre-processed count matrix in.featherformat. This should be the same normalized matrix used as input tomav_screen().
Region filtering behavior:
- Regions with
hypervar ≤ 1are automatically excluded as they represent regions with expected or reduced variance (not informative for downstream analysis) - The stratified binning ensures balanced selection across expression levels, preventing bias toward highly expressed regions
- The final
log2mean_quantile_thresthreshold removes lowly expressed regions that may be dominated by technical noise
- Regions with
Parameter tuning recommendations: If your analysis yields too few selected regions, this typically occurs when you’re analyzing a targeted genomic subset (e.g., promoter regions, CpG islands, specific gene loci) rather than genome-wide bins. The combination of default
keep_percent = 0.01(1%) andlog2mean_quantile_thres = 0.99(99th percentile) is too stringen.Scenario keep_percentlog2mean_quantile_thresExpected output Genome-wide bins (default) 0.01 (1%) 0.99 (99th) ~1,000-5,000 regions Targeted regions (promoters, peaks) 0.05-0.10 (5-10%) 0.90-0.95 (90-95th) ~2,000-10,000 regions Very sparse data 0.10-0.20 (10-20%) 0.85-0.90 (85-90th) ~5,000-20,000 regions Large datasets (>100K input regions) Reduce n_binsto 50Keep defaults Faster computation
Output Files
The function generates the following output files in the specified
out_dir (default: “./”):
- Filtered Count Matrix -
<input_name>_filtered_regions.feather- Feather format file containing count data for filtered regions
- First column “pos” contains region identifiers
- Subsequent columns contain count values for each sample
- Only includes regions passing all selection criteria:
- Hypervariance > 1 (overdispersed)
- Top hypervariant features within each log2(mean) bin
- log2(mean) above the specified quantile threshold
- Dimensions: n_selected_regions × (n_samples + 1)
| pos | H3K36me3-H3K36me3 | H3K36me3-H3K4me1 | H3K27ac-H3K36me3 | H3K36me3-H3K9me2 |
|---|---|---|---|---|
| chr1_9601_10400 | 0.03135214 | 0.02224862 | 0.03659935 | 0.03001337 |
| chr1_10401_11200 | 0.02891244 | 0.01988311 | 0.03451229 | 0.02833411 |
| chr1_11201_12000 | 0.02750192 | 0.01844290 | 0.03329854 | 0.02701144 |
| chr1_12001_12800 | 0.02933155 | 0.02015533 | 0.03510177 | 0.02985421 |
| chr1_12801_13600 | 0.02684492 | 0.01791241 | 0.03166388 | 0.02622109 |
| chr1_13601_14400 | 0.02599817 | 0.01762288 | 0.03088455 | 0.02548933 |
| chr1_14401_15200 | 0.02744561 | 0.01819877 | 0.03255291 | 0.02680714 |
| chr1_15201_16000 | 0.02819945 | 0.01888922 | 0.03394107 | 0.02790365 |
- Diagnostic Plots -
<input_name>_filtered_regions.png(ifplot = TRUE)
This combined figure contains two complementary visualizations of the selection process:
- Panel A: Mean-Variance Relationship
- Scatter plot of log2(mean) vs log2(variance)
- Shows how selected regions relate to overall mean-variance distribution
- Highlights selected regions:
- Light Green points: All regions
- Light Orange points: Final selected highly-variable regions
- Panel B: Hypervariance vs Expression
- Scatter plot of log2(mean) vs hypervariance
- Visualizes the selection process across expression levels
- Three layers of points:
- Blue points: All regions from hypervar summary
- Red points: Regions selected from binned filtering
- Green points: Final selected highly-variable regions after quantile filtering
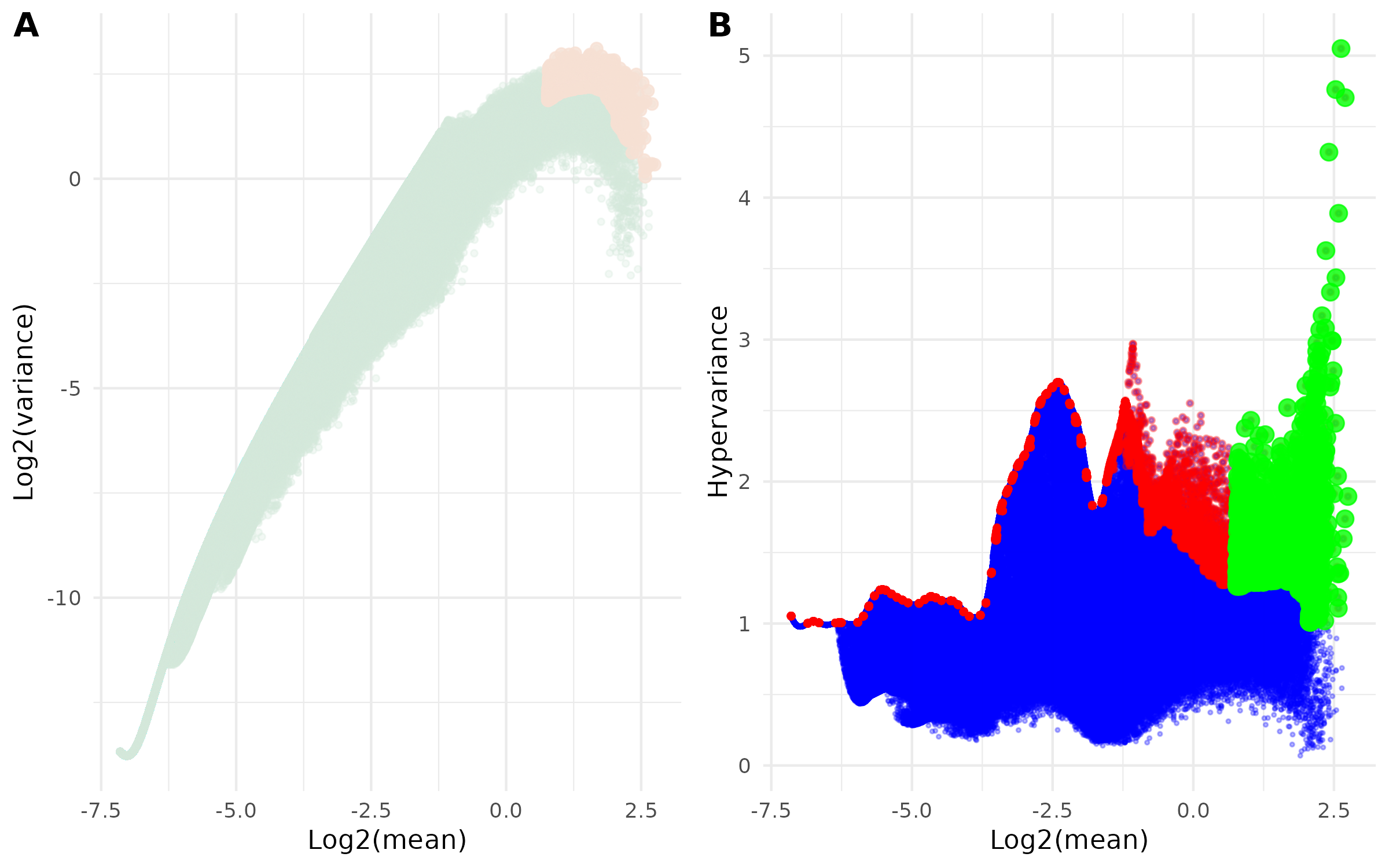
Example Usage
library(multiEpiCore)
# Test Data
trans_path <- "./count_matrix/Count_Matrix_merged_transformed.feather"
mav_path <- "bicluster/Count_Matrix_merged_transformed_mav_screen.feather"
out_dir <- "bicluster"
informative_regions(transformed_cm_pat = trans_path, mav_stats_path = mav_path, out_dir = out_dir, plot=TRUE)4. Combinatory Landscape of Co-localized CRF Pairs
4A. Automated Clustering and Heatmap Generation
The biclustering() function performs bidirectional
k-means clustering on genomic count matrices and generates cluster
assignment files along with publication-ready heatmap
visualizations.
What this function does:
(Consensus k-means clustering) Applies k-means clustering independently to rows (genomic regions) and columns (CRF pairs). For robustness, consensus clustering aggregates results from multiple k-means runs (controlled by
row_repeatsandcol_repeatsin the underlying algorithm) to identify stable cluster assignments.(Hierarchical cluster ordering) After initial k-means assignment, clusters are reordered hierarchically to optimize visual interpretation in heatmaps. For each dimension (rows/columns), cluster centroids (mean profiles) are calculated and hierarchically clustered using specified distance metrics and linkage methods. The resulting dendrogram is reordered by centroid weights to place similar clusters adjacent to each other.
(Within-cluster feature ordering) Within each cluster, individual features are reordered using hierarchical clustering to reveal internal structure and gradual transitions between expression patterns. This two-level organization (between-cluster + within-cluster) ensures both global pattern recognition and local detail preservation.
(Integrated heatmap generation) Automatically creates publication-ready heatmaps with the clustered and ordered matrix, using the generated cluster assignment files to add annotation tracks. Heatmap aesthetics (color ranges, font sizes, column name display) are fully customizable.
Parameters
| Parameter | Type | Default | Description | Example |
|---|---|---|---|---|
cm_path
|
character | — |
Path to the count matrix file in .feather format. The file
must contain a pos column, where rows represent genomic
positions and columns represent samples.
|
cm_path = “normalized_counts.feather”
|
row_km
|
integer | — | Number of k-means clusters applied to rows (genomic regions). |
row_km = 15
|
col_km
|
integer | — | Number of k-means clusters applied to columns (CFR pairs). |
col_km = 16
|
out_dir
|
character | — | Output directory used to save cluster assignment files and heatmap visualizations. |
out_dir = “./clustering_results”
|
seed
|
integer |
123
|
Random seed used to ensure reproducible k-means clustering results. |
seed = 42
|
plot
|
logical |
TRUE
|
Whether to generate and save the heatmap visualization. |
plot = FALSE
|
show_column_names
|
logical |
FALSE
|
Whether to display column names at the bottom of the heatmap. |
show_column_names = TRUE
|
lower_range
|
numeric |
NULL
|
Lower bound of the heatmap color scale. If NULL, the
minimum value from the data is used.
|
lower_range = 0
|
upper_range
|
numeric |
NULL
|
Upper bound of the heatmap color scale. If NULL, the
maximum value from the data is used.
|
upper_range = 10
|
row_title_fontsize
|
numeric |
NULL
|
Font size for row cluster titles (e.g. A, B, C). |
row_title_fontsize = 40
|
col_title_fontsize
|
numeric |
NULL
|
Font size for column cluster titles (e.g. 1, 2, 3). |
col_title_fontsize = 22
|
legend_title_fontsize
|
numeric |
NULL
|
Font size used for the heatmap legend title. |
legend_title_fontsize = 40
|
legend_label_fontsize
|
numeric |
NULL
|
Font size used for legend tick labels. |
legend_label_fontsize = 30
|
Input requirements:
- Count matrix must be in .feather format
- First column must be named “pos” containing region identifiers
- Matrix must contain numeric values only
Reproducibility: Always set
seedparameter for reproducible results, as k-means involves random initialization.Color scale interpretation:
- Default: Auto-scales to data range
- Custom: Use
lower_rangeandupper_rangefor consistent scales across analyses - Blue = Low signal, White = Medium, Red = High signal
Output Files
The function generates the following output files in the specified
out_dir:
- Row cluster assignments -
row_table.tsv- Tab-separated file with row cluster assignments
- Two columns:
region: Genomic region identifier (e.g., “chr1_1000_2000”)cluster: Cluster label as letter (A, B, C, D, etc.)
- Sorted by cluster label for easy inspection
- Rows within each cluster are ordered by hierarchical clustering
- Used for annotating genomic regions by cluster membership
region cluster <chr> <chr> 1 chr16_89995201_89996000 A 2 chr3_10401_11200 A 3 chrX_156029601_156030400 A 4 chr21_8234401_8235200 A 5 chr7_64849601_64850400 A 6 chr2_85993601_85994400 A 7 chr7_106568801_106569600 B 8 chr19_4374401_4375200 B 9 chr10_72320801_72321600 B 10 chr1_30768801_30769600 B 11 chr1_28259201_28260000 C 12 chr16_1379201_1380000 C 13 chr6_43771201_43772000 C 14 chr16_28824001_28824800 C 15 chr11_65497601_65498400 C 16 chr6_31958401_31959200 D 17 chr18_3448001_3448800 D 18 chr19_1044801_1045600 D 19 chr17_75046401_75047200 D 20 chr2_85602401_85603200 D
- Column cluster assignments -
col_table.tsv- Tab-separated file with column (sample) cluster assignments
- Two columns:
pair: CFR pair identifier (e.g., “YY1-cJun”)cluster: Cluster label as number (1, 2, 3, etc.)
- Sorted by cluster label
- Columns within each cluster are ordered by hierarchical clustering
- Used for grouping samples by similarity
pair cluster <chr> <dbl> 1 H3K9me3-H3K9me3 1 2 H3K27ac-H3K4me3 2 3 H3K4me1-H3K9me3 2 4 H3K9me2-H3K9me3 2 5 H3K27ac-H3K4me1 2 6 H3K27ac-H3K27me3 2 7 H3K27ac-H3K9me3 2 8 H3K27ac-H3K27ac 2 9 H3K4me1-H3K4me3 3 10 H3K27me3-H3K4me3 3 11 H3K4me3-H3K9me3 3 12 H3K4me3-H3K4me3 4
- Clustered heatmap -
figures/biclustering_heatmap.pdf- Publication-ready heatmap in PDF format
- Features:
- Rows organized by k-means clusters (labeled A, B, C, etc.)
- Columns organized by k-means clusters (labeled 1, 2, 3, etc.)
- Color scale: Blue (low) → White (middle) → Red (high)
- Horizontal legend positioned at bottom
- White gaps between clusters for visual separation (3mm)
- Customizable font sizes for titles and labels
- Transparent background for publication flexibility
- Dimensions automatically calculated based on matrix size (5mm per feature/sample)
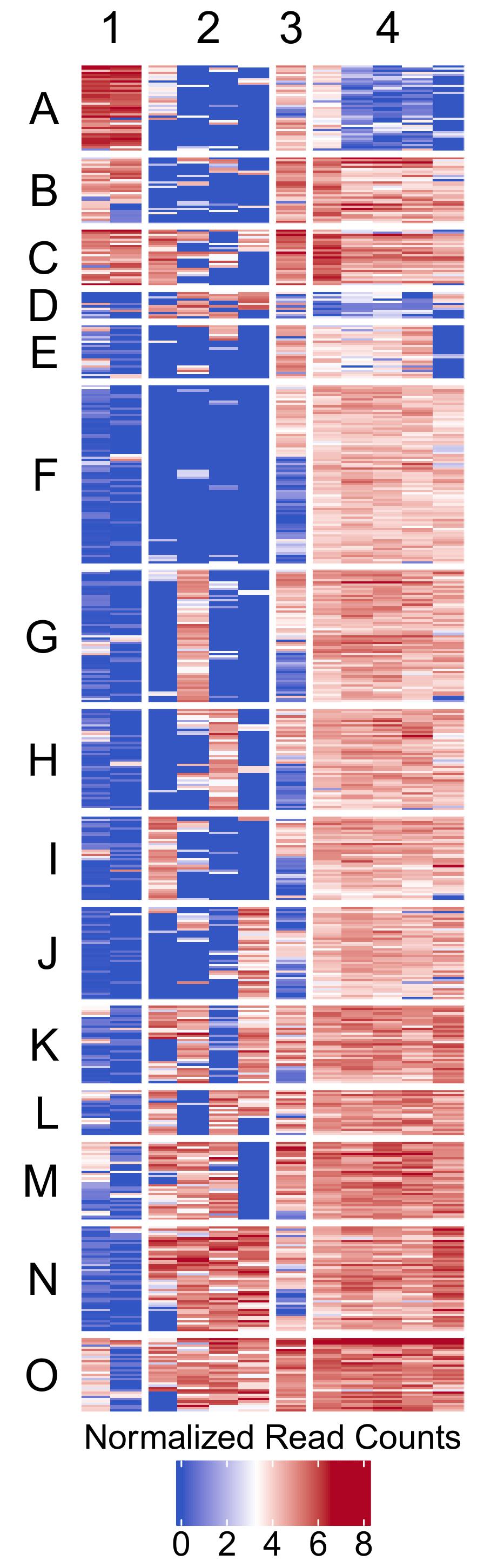
Example Usage
library(multiEpiCore)
# General Usage
cm_path <- "Count_Matrix_merged_transformed_mav_screen_filtered_regions.feather"
out_dir <- "."
biclustering(cm_path = cm_path, row_km = 15, col_km = 16, out_dir = out_dir)
# Test Data
cm_path <- "bicluster/Count_Matrix_merged_transformed_filtered_regions.feather"
out_dir <- "bicluster"
biclustering(cm_path = cm_path, row_km = 15, col_km = 4, out_dir = out_dir)4B. (Optional) Add Non-Informative Regions Back to Clusters
The add_regions_back_to_cluster() function assigns
cluster labels to genomic regions that were excluded from the highly
variable set by correlating them with existing cluster signatures.
What this function does:
This function recovers regions that were filtered out during
highly-variability selection and assigns them to the row clusters
generated by the biclustering() function.
(Filter non-highly-variable regions) Starting with the original count matrix, the function identifies non-highly-variable regions (those excluded from clustering) and filters them based on a minimum non-zero count threshold at the raw count level. Specifically, regions must have non-zero counts in more than a specified number of samples (controlled by
cutoff_non_zero) to be considered for cluster assignment.(Correlation-based cluster assignment) For each row cluster from biclustering, the function calculates a signature profile by averaging the transformed expression values of highly variable regions assigned to that cluster. Non-highly-variable regions are then correlated against these cluster signatures using their transformed values. Regions with maximum correlation values exceeding the specified quantile threshold (controlled by
quantile_threshold) are assigned to their best-matching cluster.(Priority-based label assignment) The final output combines all regions into a priority-based classification:
- Original cluster assignments from biclustering (highest priority)
- Correlation-based assignments for high-correlation regions
- CRF_specific for low-correlation candidates that passed the non-zero filter
- Background for all remaining regions with non-zero counts
Parameters
| Parameter | Type | Default | Description | Example |
|---|---|---|---|---|
orig_cm_path
|
character | — |
Path to a .feather file containing the original count
matrix generated by build_count_matrix. The matrix includes
all regions in untransformed form, with pos as the first
column.
|
orig_cm_path = “count_matrix.feather”
|
transformed_cm_path
|
character | — |
Path to a .feather file containing transformed count data
(e.g. log-normalized or scaled) used for correlation analysis.
|
transformed_cm_path = “qnorm_counts.feather”
|
filtered_cm_path
|
character | — |
Path to a .feather file containing informative or
significant regions used in the original clustering analysis.
|
filtered_cm_path = “informative_regions.feather”
|
row_cluster_path
|
character | — |
Path to a TSV file defining cluster assignments for informative regions.
The file must include region and cluster
columns.
|
row_cluster_path = “row_table.tsv”
|
out_dir
|
character | — | Output directory used to save result tables and optional diagnostic plots. |
out_dir = “./results”
|
cutoff_non_zero
|
integer |
10
|
Minimum number of non-zero samples required per region. Regions with more than this number of non-zero values are retained. |
cutoff_non_zero = 15
|
quantile_threshold
|
numeric |
0.75
|
Quantile threshold (range: 0–1) for filtering high-correlation regions. Only regions above this quantile are assigned cluster labels. |
quantile_threshold = 0.80
|
plot
|
logical |
FALSE
|
Whether to generate and save a histogram of the correlation distribution. |
plot = TRUE
|
Input file requirements:
- All feather files must have ‘pos’ column as first column
cluster_pathTSV must have ‘region’ and ‘cluster’ columns
Non-zero filtering: The
cutoff_non_zero = 10means regions must have MORE than 10 non-zero samples (not equal to 10).Quantile threshold interpretation:
quantile_threshold = 0.75means only regions with correlation in the top 25% receive cluster assignments.
Output Files
The function generates the following output files in
out_dir:
- Complete region-cluster table -
row_table_all.tsv- Comprehensive table containing all genomic regions with assigned labels
- Two columns:
region: Genomic region identifier (chromosome coordinates)cluster: Assigned cluster label or category
- Label priority hierarchy (highest to lowest):
- Original cluster assignments: From
row_cluster_path(informative region clusters) - Correlation-based assignments: High-correlation regions matched to cluster signatures
- CRF_specific: Regions passing non-zero filter but below correlation threshold
- Background: All other regions with non-zero counts
- Original cluster assignments: From
- Tab-delimited format (.tsv)
- Includes all non-zero regions from the genome
region cluster chr1_9601_10400 J chr1_10401_11200 CRF_specific chr1_12801_13600 Background chr1_14401_15200 Background chr1_15201_16000 Background chr1_16001_16800 Background - Filtered region-cluster table -
row_table_clean.tsv- Contains only cluster-assigned regions (excludes “Background” and “CRF_specific” labels)
- Same two-column format as complete table
- Tab-delimited format (.tsv)
- Useful for downstream analyses focusing on clustered regions only
- Correlation distribution histogram -
correlation_histogram.png(ifplot = TRUE)- PNG format visualization showing distribution of maximum correlation values
- X-axis: Maximum correlation between regions and cluster signatures
- Y-axis: Frequency of regions
- Red dashed line: Quantile threshold for filtering
- Legend: Shows quantile threshold value
- Resolution: 800 × 600 pixels
- Helps visualize correlation quality and threshold selection
- Color scheme: Light blue bars with black borders
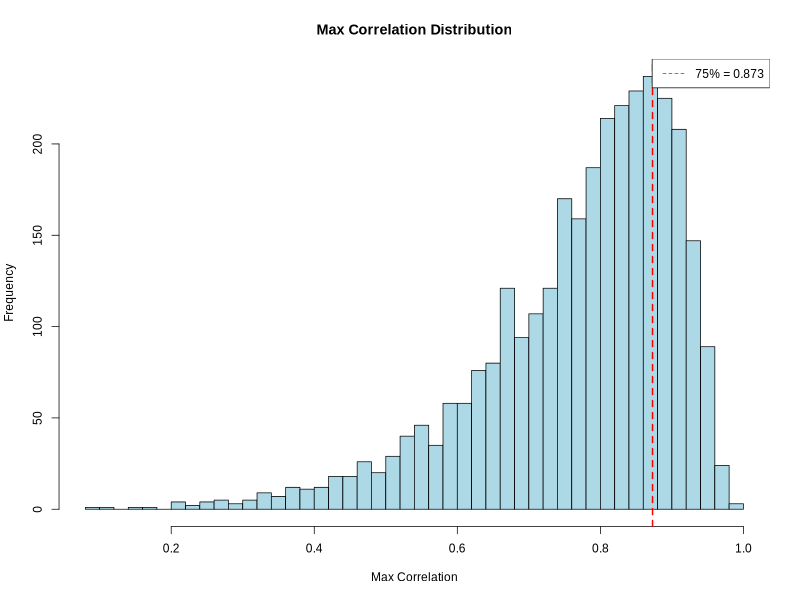
Example Usage
library(multiEpiCore)
# Test Data
orig_cm_path <- "count_matrix/Count_Matrix_merged.feather"
transformed_cm_path <- "bicluster/Count_Matrix_merged_transformed.feather"
filtered_cm_path <- "bicluster/Count_Matrix_merged_transformed_filtered_regions.feather"
row_cluster_path <- "bicluster/row_table.tsv"
out_dir <- "add_regions_results"
add_regions_back_to_cluster(
orig_cm_path = orig_cm_path,
transformed_cm_path = transformed_cm_path,
filtered_cm_path = filtered_cm_path,
row_cluster_path = row_cluster_path,
out_dir = out_dir,
cutoff_non_zero = 10,
quantile_threshold = 0.75,
plot = TRUE
)4C. (Optional) Apply Pre-Computed Clusters to Generate Heatmaps
After adding non-highly-variable regions back to clusters, the
biclustering_heatmap() function can be used to generate
heatmaps using the expanded cluster assignments (typically from
row_table_clean.tsv). This function creates
publication-ready visualizations without re-running the clustering
algorithm. Note that this function is also called internally by the
biclustering() function to generate the initial heatmap
after performing bidirectional k-means clustering.
What this function does:
(Load and validate cluster assignments) Reads row and column cluster assignment files and validates that regions in the cluster files match those present in the input count matrix. Typically uses the clean cluster assignments (
row_table_clean.tsv) that exclude “Background” and “CRF_specific” labels.(Order matrix by clusters) Reorders the count matrix rows and columns according to cluster assignments, ensuring regions within the same cluster are grouped together for visualization.
(Configure color scaling) Sets up a diverging color scheme (blue → white → red) with customizable value ranges. If ranges are not specified, automatically determines appropriate bounds from the data.
(Calculate optimal dimensions) Automatically computes cell sizes and heatmap dimensions based on the number of clusters and their sizes, ensuring cluster labels are readable and properly positioned. Adjusts legend font sizes to fit within the available space.
(Generate publication-ready heatmap) Creates a PDF heatmap with:
- Rows split by cluster assignments (labeled A, B, C, …)
- Columns split by cluster assignments (labeled 1, 2, 3, …)
- White gaps between clusters for visual separation
- Horizontal legend at bottom showing color scale
- Transparent background for publication flexibility
- High-resolution rasterized cells for efficient rendering
Parameters
| Parameter | Type | Default | Description | Example |
|---|---|---|---|---|
mat
|
matrix | — | Count matrix with genomic regions as rows and samples as columns. The matrix must contain row names (region IDs) and column names (sample IDs). |
mat = as.matrix(count_data)
|
row_cluster_file_path
|
character | — |
Path to a TSV file defining row cluster assignments. The file must
contain region and cluster columns. Typically
uses row_table_clean.tsv generated by
add_regions_back_to_cluster().
|
row_cluster_file_path = “row_table_clean.tsv”
|
col_cluster_file_path
|
character | — |
Path to a TSV file defining column cluster assignments. The file must
contain pair and cluster columns.
|
col_cluster_file_path = “col_table.tsv”
|
out_dir
|
character |
“./”
|
Output directory used to save the generated heatmap. |
out_dir = “./heatmaps”
|
show_column_names
|
logical |
FALSE
|
Whether to display sample names along the column axis of the heatmap. |
show_column_names = TRUE
|
lower_range
|
numeric |
NULL
|
Lower bound for the heatmap color scale. If NULL, the
minimum value in the matrix is used.
|
lower_range = 0
|
upper_range
|
numeric |
NULL
|
Upper bound for the heatmap color scale. If NULL, the
maximum value in the matrix is used.
|
upper_range = 4.5
|
row_title_fontsize
|
numeric |
NULL
|
Font size for row cluster titles (e.g. A, B, C). If NULL, a
default size of 20 is used.
|
row_title_fontsize = 25
|
col_title_fontsize
|
numeric |
NULL
|
Font size for column cluster titles (e.g. 1, 2, 3). If
NULL, a default size of 20 is used.
|
col_title_fontsize = 25
|
legend_title_fontsize
|
numeric |
NULL
|
Font size for the legend title. If NULL, a default size of
15 is used and auto-adjusted to fit.
|
legend_title_fontsize = 18
|
legend_label_fontsize
|
numeric |
NULL
|
Font size for legend tick labels. If NULL, a default size
of 15 is used.
|
legend_label_fontsize = 15
|
Note: The input mat is typically the
complete transformed count matrix (e.g., from
transformed_cm_path). The function will automatically
subset the matrix to include only regions present in both the matrix and
the cluster assignment files. Regions in the matrix but not in cluster
files will be excluded from visualization; regions in cluster files but
not in the matrix will be skipped.
Output Files
The function generates the following output files in the specified
out_dir:
- Clustered heatmap(s) -
biclustering_heatmap.pdf- Publication-ready heatmap(s) in PDF format
- One PDF file per input count matrix
- Features:
- Rows ordered and split by cluster assignments from
row_cluster_file_path - Columns ordered and split by cluster assignments from
col_cluster_file_path - Row clusters labeled with letters (A, B, C, etc.)
- Column clusters labeled with numbers (1, 2, 3, etc.)
- Color scale: Blue (#3155C3) → White → Red (#AF0525)
- Horizontal legend positioned at bottom with title “Normalized Read Counts”
- White gaps between clusters (3mm) for visual separation
- Transparent background for publication flexibility
- No dendrograms (unless show_dend_boolean = TRUE shows column names)
- Rows ordered and split by cluster assignments from
- Dimensions automatically calculated based on matrix size (5mm per feature/sample)
Example Usage
library(multiEpiCore)
# Add non-highly-variable regions back to clusters
orig_cm_path <- "./count_matrix.feather"
transformed_cm_path <- "./count_matrix_log2_qnorm.feather"
filtered_cm_path <- "./highly_variable_regions.feather"
row_cluster_path <- "./biclustering_results/row_table.tsv"
out_dir <- "./add_regions_results"
add_regions_back_to_cluster(
orig_cm_path = orig_cm_path,
transformed_cm_path = transformed_cm_path,
filtered_cm_path = filtered_cm_path,
row_cluster_path = row_cluster_path,
out_dir = out_dir,
cutoff_non_zero = 10,
quantile_threshold = 0.75,
plot = TRUE
)
# Load the transformed count matrix
library(arrow)
library(tibble)
mat <- as.matrix(column_to_rownames(read_feather(transformed_cm_path), var = "pos"))
# Generate heatmap with expanded cluster assignments
row_table_clean_path <- file.path(out_dir, "row_table_clean.tsv")
col_table_path <- "./biclustering_results/col_table.tsv"
biclustering_heatmap(
mat = mat,
row_cluster_file_path = row_table_clean_path,
col_cluster_file_path = col_table_path,
out_dir = out_dir
)5. (Optional) Cluster-based Annotation
5A. Annotate and Plot Regulatory Element Composition for Clustered Regions
The biclustering_genomic_distribution() function
performs post-clustering genomic annotation analysis by quantifying the
regulatory element composition of clustered genomic regions. Clustered
regions are overlapped with multiple external annotation resources,
including cCREs, ChromHMM chromatin states, and repeat elements,
followed by comparative visualization across clusters.
Parameters
| Parameter | Type | Default | Description | Example |
|---|---|---|---|---|
row_cluster_file_path
|
character | — |
Path to a TSV file containing cluster assignments. The file must include
region (genomic coordinates formatted as chr_start_end) and
cluster (cluster ID) columns. Typically uses
row_table_clean.tsv generated from biclustering output.
|
“./row_table_clean.tsv”
|
out_dir
|
character |
“./”
|
Output directory for annotation results. Subdirectories are automatically created for each annotation type. |
“./distribution_annotation”
|
distributions
|
character vector |
c(“genic”, “ccre”)
|
Annotation types to perform. Supported options include
“genic” (gene features), “ccre” (cCRE
elements), “chromhmm” (chromatin states), and
“repeat” (repeat elements). Any combination of these
options can be specified.
|
c(“genic”, “ccre”, “repeat”)
|
ref_genome
|
character |
“hg38”
|
Reference genome version. Supported options are “hg38”
(Human GRCh38) and “mm10” (Mouse GRCm38).
|
“mm10”
|
ref_source
|
character |
“knownGene”
|
Gene annotation source used for genic and cCRE annotation. Supported
options are “knownGene” (UCSC) and “GENCODE”.
This parameter is only used when “genic” is included in
distributions.
|
“GENCODE”
|
mode
|
character |
“nearest”
|
Annotation assignment method. “nearest” assigns each region
to the closest feature, while “weighted” assigns features
proportionally based on overlap length.
|
“weighted”
|
plot
|
logical |
TRUE
|
Whether to generate stacked barplot visualizations for each annotation type. |
FALSE
|
- Annotation types available:
"genic": Gene structural features - Promoter, 5’ UTR, Exon, Intron, 3’ UTR"ccre": Candidate cis-Regulatory Elements - dELS, pELS, PLS, CA-H3K4me3, CA-CTCF, CA-TF, TF, CA"chromhmm": Chromatin states - Acet, EnhWk, EnhA, PromF, TSS, TxWk, TxEx, Tx, OpenC, TxEnh, ReprPCopenC, BivProm, ZNF, ReprPC, HET, GapArtf, Quies"repeat": Repetitive elements - SINE, LINE, LTR, Retroposon, RC, DNA, Satellite, Simple_repeat, Low_complexity, rRNA, tRNA, snRNA, scRNA, srpRNA, RNA, Unknown- For detailed description of each annotation category, see the Annotation page
- Annotation assignment methods:
"nearest": Each genomic region is assigned to its closest feature (by distance to feature center)"weighted": Each region is proportionally assigned to overlapping features based on overlap lengthnearestmode is faster and simpler;weightedmode provides more accurate representation for regions spanning multiple features
Output Files
The function generates the following output files in the specified
out_dir:
- Composition tables -
{genic/ccre/chromhmm/repeat}_distribution.pdf
.tsv tables summarizing the percentage composition of various annotations
Row: cluster lables
Col: annotations states
Values represent the proportion (%) of genomic regions assigned to each state within a cluster
Genic distribution
Promoter
5’ UTR
Exon
Intron
3’ UTR
other
A
7.37327188940092
0
2.76497695852535
13.3640552995392
0
76.4976958525346
B
59.8802395209581
11.9760479041916
8.98203592814371
16.7664670658683
1.79640718562874
0.598802395209581
C
54.2253521126761
11.9718309859155
13.3802816901408
14.0845070422535
0
6.33802816901408
D
8.82352941176471
0
10.2941176470588
54.4117647058823
2.94117647058824
23.5294117647059
E
43.3823529411765
10.2941176470588
10.2941176470588
27.2058823529412
1.47058823529412
7.35294117647059
……
O
60.427807486631
10.6951871657754
13.903743315508
12.8342245989305
0
2.13903743315508
cCRE distribution
dELS
pELS
PLS
CA-H3K4me3
A
26.7281105990783
7.37327188940092
2.76497695852535
9.67741935483871
B
5.38922155688623
34.7305389221557
59.8802395209581
0
C
4.22535211267606
26.7605633802817
62.6760563380282
3.52112676056338
D
79.4117647058823
13.2352941176471
2.94117647058824
0
E
19.8529411764706
41.9117647058824
38.2352941176471
0
……
O
4.81283422459893
28.8770053475936
66.3101604278075
0
CA-CTCF
CA-TF
TF
CA
other
A
0.921658986175115
0
4.14746543778802
1.84331797235023
46.5437788018433
B
0
0
0
0
0
C
0
0
1.40845070422535
0
1.40845070422535
D
0
0
2.94117647058824
0
1.47058823529412
E
0
0
0
0
0
……
O
0
0
0
0
0
chromHMM distribution
Acet
EnhWk
EnhA
PromF
TSS
OpenC
A
3.2258064516129
0.921658986175115
2.76497695852535
5.06912442396313
0.921658986175115
0
B
0
0
0.598802395209581
43.7125748502994
50.8982035928144
0.598802395209581
C
1.40845070422535
0
1.40845070422535
50.7042253521127
35.9154929577465
2.11267605633803
D
16.1764705882353
0
36.7647058823529
16.1764705882353
0
2.94117647058824
E
1.47058823529412
0.735294117647059
4.41176470588235
38.2352941176471
38.2352941176471
2.20588235294118
……
O
0.53475935828877
0
2.67379679144385
45.4545454545455
49.7326203208556
0
TxEnh
BivProm
TxWk
TxEx
Tx
A
0.921658986175115
0.921658986175115
0
0
0
B
0
1.19760479041916
0
0
0
C
0.704225352112676
0
0
0
0
D
11.7647058823529
0
1.47058823529412
10.2941176470588
1.47058823529412
E
0.735294117647059
10.2941176470588
0
0
0
……
O
0
1.06951871657754
0
0
0
ZNF
ReprPC
HET
GapArtf
Quies
other
A
0.921658986175115
3.2258064516129
13.3640552995392
38.2488479262673
1.84331797235023
27.6497695852535
B
0
0
1.19760479041916
0
0
1.79640718562874
C
0
0
1.40845070422535
2.11267605633803
0
4.22535211267606
D
0
0
0
1.47058823529412
0
1.47058823529412
E
0
2.94117647058824
0.735294117647059
0
0
0
……
O
0
0
0
0
0
0.53475935828877
Repeat distribution
SINE
LINE
LTR
Retroposon
RC
DNA
A
9.21658986175115
5.52995391705069
5.06912442396313
0.460829493087558
0
0
B
10.7784431137725
5.38922155688623
8.38323353293413
0
0
1.79640718562874
C
11.2676056338028
4.22535211267606
7.04225352112676
0
0
0
D
27.9411764705882
14.7058823529412
13.2352941176471
0
0
7.35294117647059
E
10.2941176470588
6.61764705882353
5.14705882352941
0
0
2.20588235294118
……
O
8.55614973262032
4.27807486631016
1.06951871657754
0.53475935828877
0
1.60427807486631
Satellite
Simple_repeat
Low_complexity
rRNA
tRNA
snRNA
A
36.405529953917
25.3456221198157
0.921658986175115
0
0
0
B
1.19760479041916
29.3413173652695
8.98203592814371
0
1.19760479041916
0
C
2.11267605633803
30.2816901408451
7.04225352112676
0
0
0
D
2.94117647058824
7.35294117647059
1.47058823529412
0
0
0
E
0
33.8235294117647
14.7058823529412
0
0
0
O
0
36.3636363636364
9.62566844919786
0
0.53475935828877
0
scRNA
srpRNA
RNA
Unknown
other
A
0
0
0
0
17.0506912442396
B
0
0.598802395209581
0
0
32.3353293413174
C
0
0
0
0
38.0281690140845
D
0
0
0
0
25
E
0
0
0
0
27.2058823529412
……
O
0
0
0
0
37.4331550802139
- Cmposition bar plot -
{genic/ccre/chromhmm/repeat}_distribution.pdf- Stacked horizontal bar plot showing ChromHMM chromatin state distribution across clusters
- X-axis: Percentage (0-100%)
- Y-axis: Cluster labels (top to bottom)
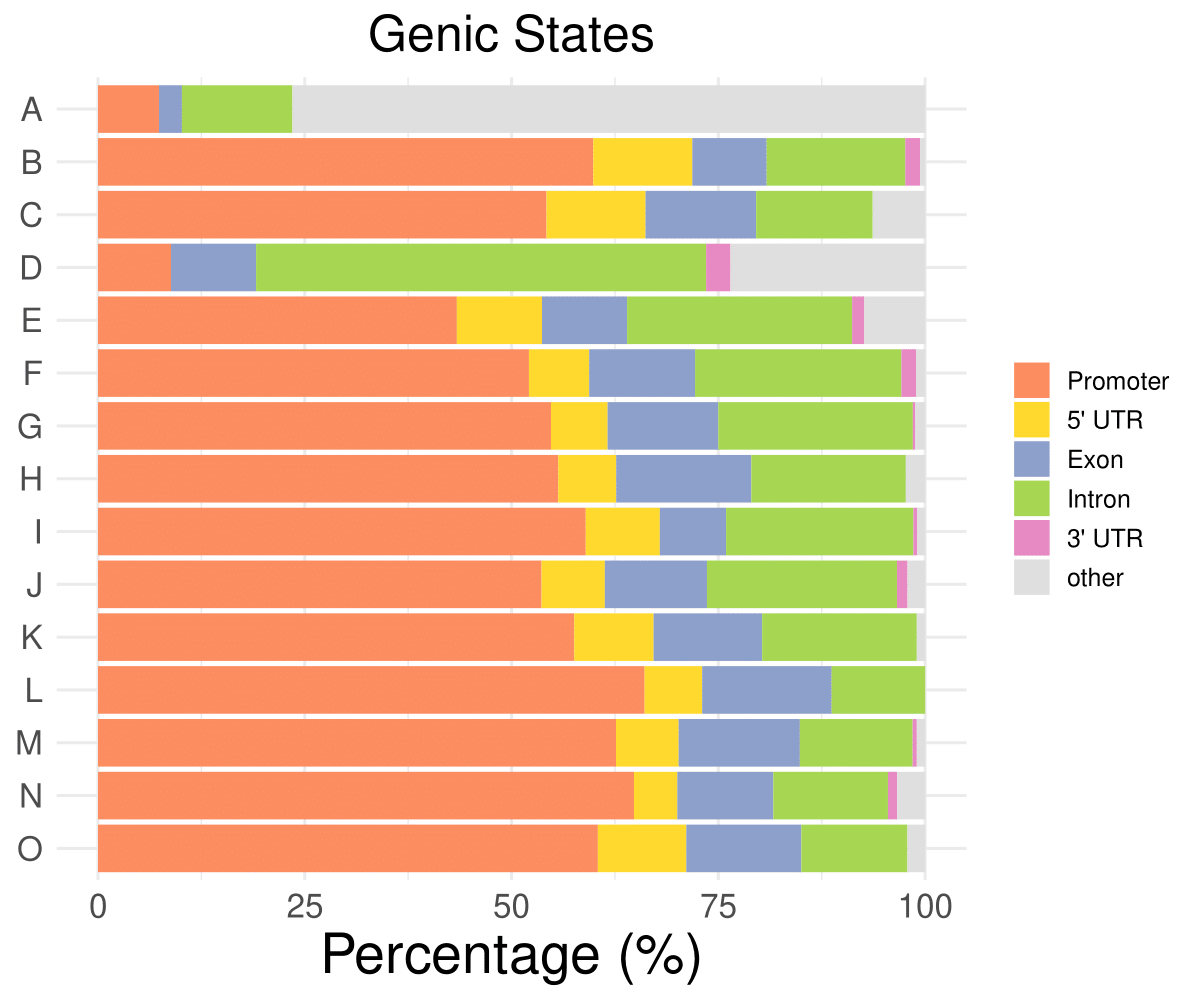
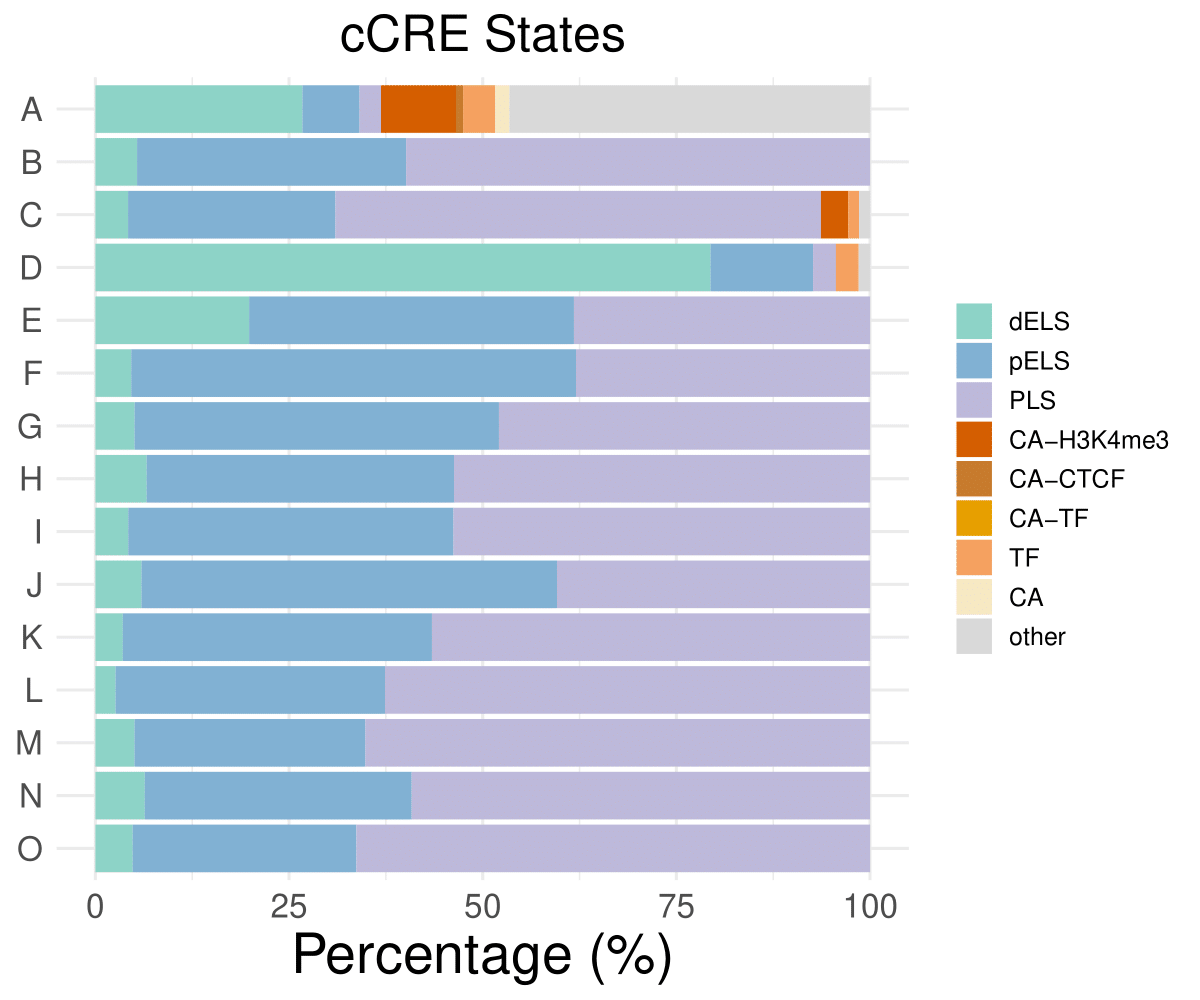
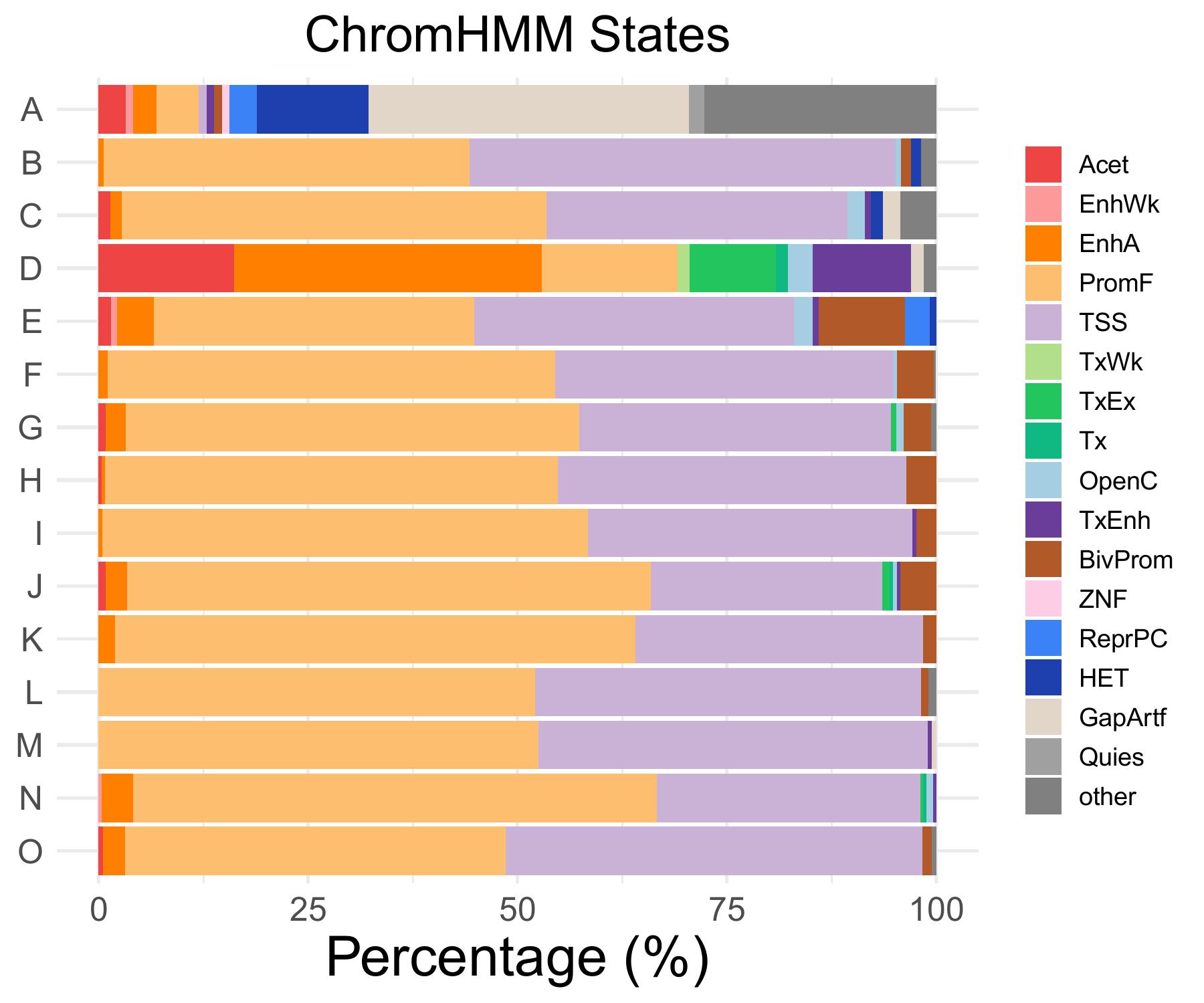
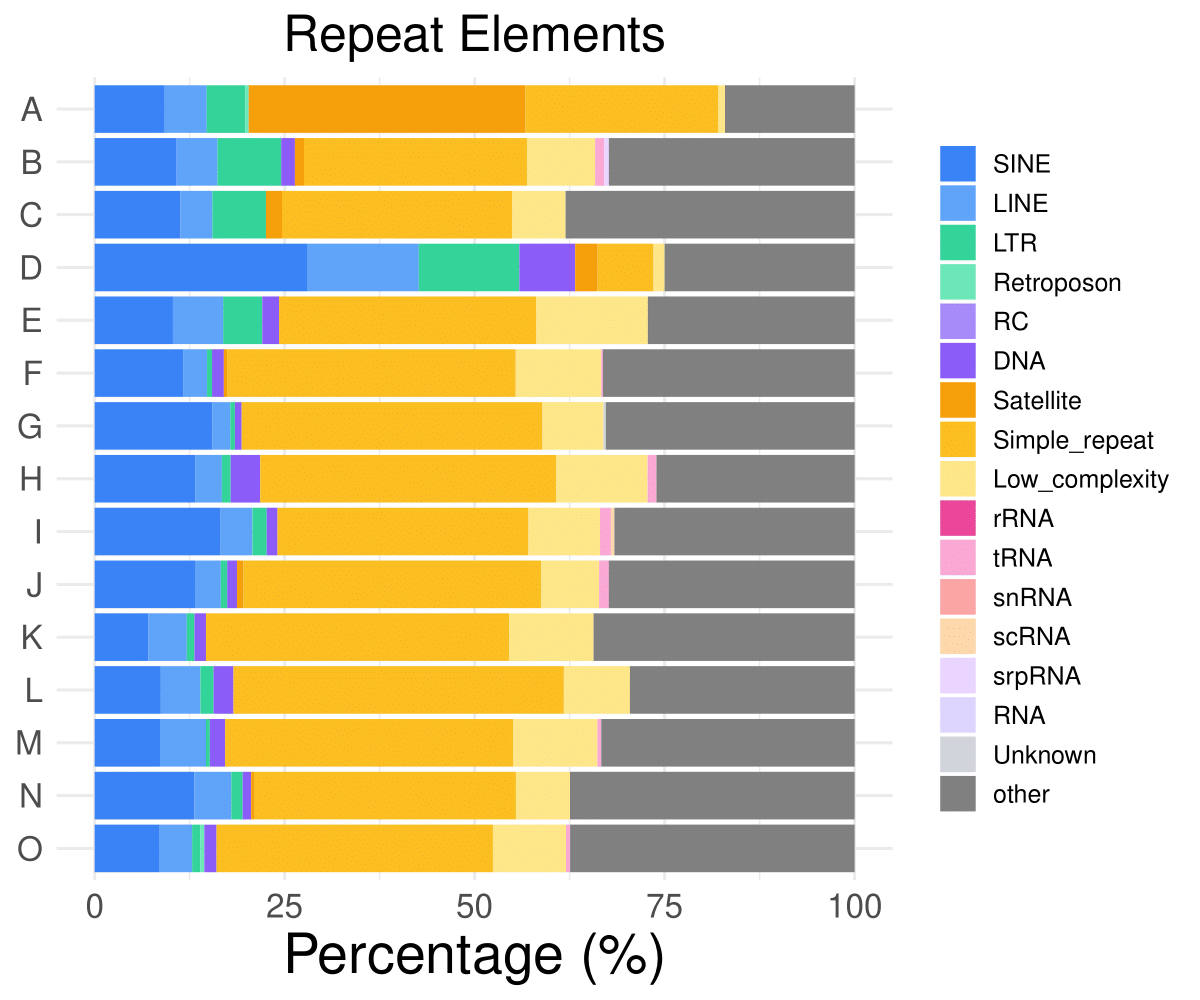
Example Usage
library(multiEpiCore)
# Test Data
row_cluster_file_path <- "bicluster/row_table.tsv"
out_dir <- "bicluster/genomic_distribution"
annotation_ccre_hmm(row_cluster_file_path = row_cluster_file_path, out_dir = out_dir)5B. Biclustering TFBS Annotation Pipeline
The biclustering_TFBS_enrichment() function provides a
complete, automated workflow for analyzing transcription factor binding
site (TFBS) enrichment across multiple clusters of genomic regions. It
handles the entire pipeline from control region generation to enrichment
testing and visualization.
What this function does:
Reads a file containing clustered genomic regions (e.g., from biclustering analysis)
Generates matched control regions for each cluster using gene-based matching
Performs TFBS enrichment analysis comparing each cluster against controls
Creates heatmap visualizations showing enrichment patterns across clusters
Saves all intermediate and final results to organized output files
Parameters
| Parameter | Type | Default | Description | Example |
|---|---|---|---|---|
row_cluster_file_path
|
character | — |
Path to a tab-delimited file containing clustered regions. The file must
include columns region (formatted as chr_start_end) and
cluster (cluster ID).
|
“bicluster_results.tsv”
|
out_dir
|
character |
“./”
|
Output directory where all results will be saved. |
out_dir = “./TFBS_results/”
|
ref_genome
|
character |
“hg38”
|
Reference genome version. Supported options are “hg38”,
“hg19”, “mm10”, and “mm39”.
|
ref_genome = “mm10”
|
ref_source
|
character |
“knownGene”
|
Gene annotation source used for control region generation. Supported
options are “knownGene” (UCSC knownGene) and
“GENCODE” (GENCODE gene models).
|
ref_source = “GENCODE”
|
control_rep
|
integer |
1
|
Multiplier for control region generation, defining the ratio of control
regions to target regions. For example, setting control_rep =
2 generates twice as many control regions.
|
control_rep = 3
|
regions
|
integer |
800
|
Size in base pairs to which all regions are resized, centered on the original region midpoint. |
regions = 500
|
plot
|
logical |
TRUE
|
Whether to generate heatmap visualizations. If set to
FALSE, only enrichment analysis is performed.
|
plot = FALSE
|
Input File Format:
The cluster file must be tab-delimited with at least two columns: -
region: Genomic coordinates in format “chr_start_end”
(underscore-separated) - cluster: Cluster assignment (e.g.,
“cluster_1”, “group_A”, “bicluster_2”)
Output Files
All output files are saved to the specified out_dir:
- Matched control regions -
all_controls.bed- BED file containing all matched control regions used as background for TFBS enrichment analysis
- Regions are combined across clusters and de-duplicated to avoid redundant counting
| chr | start | end | |
|---|---|---|---|
| 1 | chr21 | 6497621 | 6498421 |
| 2 | chr21 | 8197426 | 8198226 |
| 3 | chr21 | 10482659 | 10483459 |
| 4 | chr21 | 13979717 | 13980517 |
| 5 | chr21 | 14216386 | 14217186 |
| 6 | chr21 | 15729512 | 15730312 |
| 7 | chr21 | 16301884 | 16302684 |
| 8 | chr21 | 25735402 | 25736202 |
| 9 | chr21 | 26059459 | 26060259 |
- Cluster-level TFBS enrichment table -
TFBS_enrichment_cluster_<label>.tsv
- Per-cluster transcription factor binding site enrichment results
- Includes odds ratio, p-value, FDR, and hit counts for each TF motif
| feature | target_hit | control_hit | target_off | control_off | odds_ratio | pvalue | odds_ratio_se | FDR |
|---|---|---|---|---|---|---|---|---|
| CREBBP | 11 | 4 | 206 | 3086 | 35.686987900689 | 2.41692888678869e-11 | 0.537110966474507 | 7.46831026017705e-09 |
| NCOA2 | 10 | 22 | 207 | 3068 | 7.04766662951468 | 6.09470440920052e-06 | 0.373524943105363 | 0.000812607561274904 |
| STAT5A | 12 | 34 | 205 | 3056 | 5.50587515453413 | 7.88939379878547e-06 | 0.332680015204062 | 0.000812607561274904 |
| MCM5 | 3 | 0 | 214 | 3090 | 57.3957662256922 | 8.84174540651768e-05 | 1.1202565253937 | 0.00546419866122793 |
| ZNF274 | 3 | 0 | 214 | 3090 | 57.3957662256922 | 8.84174540651768e-05 | 1.1202565253937 | 0.00546419866122793 |
| PHB2 | 10 | 36 | 207 | 3054 | 4.36306397950694 | 0.000210357129487557 | 0.350814969016483 | 0.0108333921686092 |
- Full TFBS enrichment heatmap -
TFBS_heatmap_all.pdf(ifplot = TRUE)- Heatmap visualization of enriched TFBS across all clusters
- Log2-transformed odds ratios with a symmetric color scale
- Rows: TFBS
- Columns: Cluster labels

- Top N TFBS heatmap -
TFBS_heatmap_top<n>.pdf(iftop_nprovided andplot = TRUE)- Heatmap showing the top N TFBS with the highest coefficient of variation across clusters
- Highlights TFBS exhibiting the greatest variability between clusters
- Uses the same log2 odds ratio color scale as the full heatmap for consistency
- Rows: TFBS
- Columns: Cluster labels
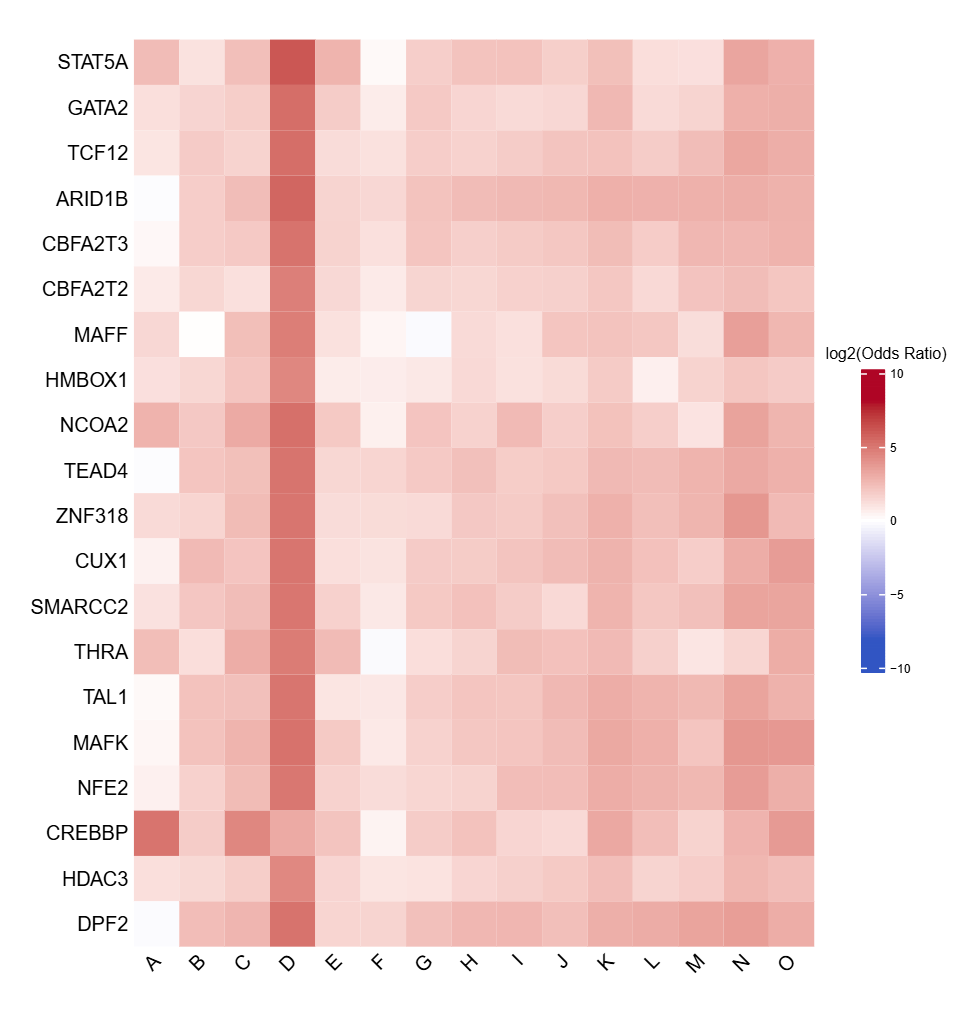
- Log2 odds ratio matrix -
odds_ratio_log2.csv(ifplot = TRUE)- Matrix of log2-transformed odds ratios for all TFBS × clusters
- Numerical data underlying the TFBS enrichment heatmap
- Rows: TFBS
- Columns: Cluster labels
- FDR matrix -
FDR.csv(ifplot = TRUE)- Matrix of FDR-adjusted p-values for all TFBS × clusters
- Same row and column order as
odds_ratio_log2.csv
Example Usage
library(multiEpiCore)
# Basic usage - complete pipeline with visualization
biclustering_TFBS_enrichment(
row_cluster_file_path = "NMF_clusters.tsv",
out_dir = "./TFBS_analysis/",
ref_genome = "hg38"
)
# Custom region size for enhancer analysis
biclustering_TFBS_enrichment(
row_cluster_file_path = "enhancer_clusters.tsv",
out_dir = "./enhancer_TFBS/",
ref_genome = "hg38",
regions = 1000
)
# Mouse genome analysis
biclustering_TFBS_enrichment(
row_cluster_file_path = "mouse_peaks_clustered.tsv",
out_dir = "./mouse_TFBS/",
ref_genome = "mm10",
regions = 500
)
# Generate 3x more control regions for more robust statistics
biclustering_TFBS_enrichment(
row_cluster_file_path = "ATAC_peaks_clustered.tsv",
out_dir = "./ATAC_TFBS/",
ref_genome = "hg38",
control_rep = 3,
regions = 800
)
# Enrichment analysis only (no heatmap)
biclustering_TFBS_enrichment(
row_cluster_file_path = "clusters.tsv",
out_dir = "./TFBS_tables/",
ref_genome = "hg38",
plot = FALSE
)
# Test Data
row_cluster_file_path <- "bicluster/row_table.tsv"
out_dir <- "bicluster/TFBS_enrichment"
annotation_ccre_hmm(row_cluster_file_path = row_cluster_file_path, out_dir = out_dir)